Evolutionary conservation analysis increases the colocalization of predicted exonic splicing enhancers in the BRCA1 gene with missense sequence changes and in-frame deletions, but not polymorphisms
- PMID: 16280041
- PMCID: PMC1410749
- DOI: 10.1186/bcr1324
Evolutionary conservation analysis increases the colocalization of predicted exonic splicing enhancers in the BRCA1 gene with missense sequence changes and in-frame deletions, but not polymorphisms
Abstract
Introduction: Aberrant pre-mRNA splicing can be more detrimental to the function of a gene than changes in the length or nature of the encoded amino acid sequence. Although predicting the effects of changes in consensus 5' and 3' splice sites near intron:exon boundaries is relatively straightforward, predicting the possible effects of changes in exonic splicing enhancers (ESEs) remains a challenge.
Methods: As an initial step toward determining which ESEs predicted by the web-based tool ESEfinder in the breast cancer susceptibility gene BRCA1 are likely to be functional, we have determined their evolutionary conservation and compared their location with known BRCA1 sequence variants.
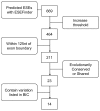
Results: Using the default settings of ESEfinder, we initially detected 669 potential ESEs in the coding region of the BRCA1 gene. Increasing the threshold score reduced the total number to 464, while taking into consideration the proximity to splice donor and acceptor sites reduced the number to 211. Approximately 11% of these ESEs (23/211) either are identical at the nucleotide level in human, primates, mouse, cow, dog and opossum Brca1 (conserved) or are detectable by ESEfinder in the same position in the Brca1 sequence (shared). The frequency of conserved and shared predicted ESEs between human and mouse is higher in BRCA1 exons (2.8 per 100 nucleotides) than in introns (0.6 per 100 nucleotides). Of conserved or shared putative ESEs, 61% (14/23) were predicted to be affected by sequence variants reported in the Breast Cancer Information Core database. Applying the filters described above increased the colocalization of predicted ESEs with missense changes, in-frame deletions and unclassified variants predicted to be deleterious to protein function, whereas they decreased the colocalization with known polymorphisms or unclassified variants predicted to be neutral.
Conclusion: In this report we show that evolutionary conservation analysis may be used to improve the specificity of an ESE prediction tool. This is the first report on the prediction of the frequency and distribution of ESEs in the BRCA1 gene, and it is the first reported attempt to predict which ESEs are most likely to be functional and therefore which sequence variants in ESEs are most likely to be pathogenic.
Figures
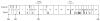
Similar articles
-
Colocalisation of predicted exonic splicing enhancers in BRCA2 with reported sequence variants.Breast Cancer Res Treat. 2008 Jul;110(2):227-34. doi: 10.1007/s10549-007-9714-5. Epub 2007 Sep 26. Breast Cancer Res Treat. 2008. PMID: 17899372
-
Missense mutations in cancer suppressor gene TP53 are colocalized with exonic splicing enhancers (ESEs).Mutat Res. 2004 Oct 4;554(1-2):175-83. doi: 10.1016/j.mrfmmm.2004.04.014. Mutat Res. 2004. PMID: 15450416
-
Unclassified variants identified in BRCA1 exon 11: Consequences on splicing.Genes Chromosomes Cancer. 2008 May;47(5):418-26. doi: 10.1002/gcc.20546. Genes Chromosomes Cancer. 2008. PMID: 18273839
-
Alterations of pre-mRNA splicing in cancer.Genes Chromosomes Cancer. 2005 Apr;42(4):342-57. doi: 10.1002/gcc.20156. Genes Chromosomes Cancer. 2005. PMID: 15648050 Review.
-
New insights into ocular albinism type 1 (OA1): Mutations and polymorphisms of the OA1 gene.Hum Mutat. 2002 Feb;19(2):85-92. doi: 10.1002/humu.10034. Hum Mutat. 2002. PMID: 11793467 Review.
Cited by
-
Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes.Breast Cancer Res. 2012 May 25;14(3):R87. doi: 10.1186/bcr3202. Breast Cancer Res. 2012. PMID: 22632462 Free PMC article.
-
Recognition of functional genetic polymorphism using ESE motif definition: a conservative evolutionary approach to CYP2D6/CYP2C19 gene variants.Genetica. 2022 Oct;150(5):289-297. doi: 10.1007/s10709-022-00161-x. Epub 2022 Aug 1. Genetica. 2022. PMID: 35913522
-
DNA damage repair system in C57BL/6 J mice is evolutionarily stable.BMC Genomics. 2021 Sep 17;22(1):669. doi: 10.1186/s12864-021-07983-7. BMC Genomics. 2021. PMID: 34535077 Free PMC article.
-
Analytical methods for inferring functional effects of single base pair substitutions in human cancers.Hum Genet. 2009 Oct;126(4):481-98. doi: 10.1007/s00439-009-0677-y. Epub 2009 May 12. Hum Genet. 2009. PMID: 19434427 Free PMC article. Review.
-
Alternative splicing and tumor progression.Curr Genomics. 2008 Dec;9(8):556-70. doi: 10.2174/138920208786847971. Curr Genomics. 2008. PMID: 19516963 Free PMC article.
References
-
- Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55–58. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

