Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei
- PMID: 16281058
- PMCID: PMC1356302
- DOI: 10.1038/sj.emboj.7600861
Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei
Abstract
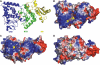
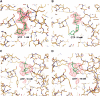
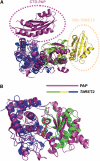
Trypanosomatids are pathogenic protozoa that undergo a unique form of post-transcriptional RNA editing that inserts or deletes uridine nucleotides in many mitochondrial pre-mRNAs. Editing is catalyzed by a large multiprotein complex, the editosome. A key editosome enzyme, RNA editing terminal uridylyl transferase 2 (TUTase 2; RET2) catalyzes the uridylate addition reaction. Here, we report the 1.8 A crystal structure of the Trypanosoma brucei RET2 apoenzyme and its complexes with uridine nucleotides. This structure reveals that the specificity of the TUTase for UTP is determined by a crucial water molecule that is exquisitely positioned by the conserved carboxylates D421 and E424 to sense a hydrogen atom on the N3 position of the uridine base. The three-domain structure also unveils a unique domain arrangement not seen before in the nucleotidyltansferase superfamily, with a large domain insertion between the catalytic aspartates. This insertion is present in all trypanosomatid TUTases. We also show that TbRET2 is essential for survival of the bloodstream form of the parasite and therefore is a potential target for drug therapy.
Figures




References
-
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte M (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 - PubMed
-
- Aphasizhev R, Aphasizheva I, Simpson L (2004) Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett 572: 15–18 - PubMed
-
- Aphasizhev R, Sbicego S, Peris M, Jang S, Aphasizheva I, Simpson A, Rivlin A, Simpson L (2002) Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108: 637–648 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

