Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco
- PMID: 16282373
- PMCID: PMC1287997
- DOI: 10.1073/pnas.0508042102
Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco
Abstract
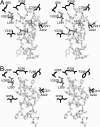
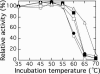
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) catalyzes the rate-limiting step of photosynthetic CO(2) fixation and, thus, limits agricultural productivity. However, Rubisco enzymes from different species have different catalytic constants. If the structural basis for such differences were known, a rationale could be developed for genetically engineering an improved enzyme. Residues at the bottom of the large-subunit alpha/beta-barrel active site of Rubisco from the green alga Chlamydomonas reinhardtii (methyl-Cys-256, Lys-258, and Ile-265) were previously changed through directed mutagenesis and chloroplast transformation to residues characteristic of land-plant Rubisco (Phe-256, Arg-258, and Val-265). The resultant enzyme has decreases in carboxylation efficiency and CO(2)/O(2) specificity, despite the fact that land-plant Rubisco has greater specificity than the Chlamydomonas enzyme. Because the residues are close to a variable loop between beta-strands A and B of the small subunit that can also affect catalysis, additional substitutions were created at this interface. When large-subunit Val-221 and Val-235 were changed to land-plant Cys-221 and Ile-235, they complemented the original substitutions and returned CO(2)/O(2) specificity to the normal level. Further substitution with the shorter betaA-betaB loop of the spinach small subunit caused a 12-17% increase in specificity. The enhanced CO(2)/O(2) specificity of the mutant enzyme is lower than that of the spinach enzyme, but the carboxylation and oxygenation kinetic constants are nearly indistinguishable from those of spinach and substantially different from those of Chlamydomonas Rubisco. Thus, this interface between large and small subunits, far from the active site, contributes significantly to the differences in catalytic properties between algal and land-plant Rubisco enzymes.
Figures



Similar articles
-
Alanine-scanning mutagenesis of the small-subunit beta A-beta B loop of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase: substitution at Arg-71 affects thermal stability and CO2/O2 specificity.Biochemistry. 2001 May 15;40(19):5615-21. doi: 10.1021/bi002943e. Biochemistry. 2001. PMID: 11341826
-
Assessment of structural and functional divergence far from the large subunit active site of ribulose-1,5-bisphosphate carboxylase/oxygenase.J Biol Chem. 2003 Dec 5;278(49):49401-5. doi: 10.1074/jbc.M309993200. Epub 2003 Sep 23. J Biol Chem. 2003. PMID: 14506244
-
Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase.Biochemistry. 2005 Jul 26;44(29):9851-61. doi: 10.1021/bi050537v. Biochemistry. 2005. PMID: 16026157
-
Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase.Arch Biochem Biophys. 2003 Jun 15;414(2):141-9. doi: 10.1016/s0003-9861(03)00171-1. Arch Biochem Biophys. 2003. PMID: 12781765 Review.
-
Improving photosynthesis through the enhancement of Rubisco carboxylation capacity.Biochem Soc Trans. 2021 Nov 1;49(5):2007-2019. doi: 10.1042/BST20201056. Biochem Soc Trans. 2021. PMID: 34623388 Review.
Cited by
-
Quantitative analysis of an engineered CO2-fixing Escherichia coli reveals great potential of heterotrophic CO2 fixation.Biotechnol Biofuels. 2015 Jun 18;8:86. doi: 10.1186/s13068-015-0268-1. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 26097503 Free PMC article.
-
The dependency of red Rubisco on its cognate activase for enhancing plant photosynthesis and growth.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25890-25896. doi: 10.1073/pnas.2011641117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989135 Free PMC article.
-
Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria.Proc Natl Acad Sci U S A. 2011 Aug 30;108(35):14688-93. doi: 10.1073/pnas.1109503108. Epub 2011 Aug 17. Proc Natl Acad Sci U S A. 2011. PMID: 21849620 Free PMC article.
-
Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate.Acta Crystallogr D Struct Biol. 2018 Jan 1;74(Pt 1):1-9. doi: 10.1107/S2059798317017132. Epub 2018 Jan 1. Acta Crystallogr D Struct Biol. 2018. PMID: 29372894 Free PMC article.
-
Temperature responses of the Rubisco maximum carboxylase activity across domains of life: phylogenetic signals, trade-offs, and importance for carbon gain.Photosynth Res. 2015 Feb;123(2):183-201. doi: 10.1007/s11120-014-0067-8. Epub 2014 Dec 17. Photosynth Res. 2015. PMID: 25515770
References
-
- Spreitzer, R. J. & Salvucci, M. E. (2002) Annu. Rev. Plant Biol. 53, 449-475. - PubMed
-
- Andersson, I. & Taylor, T. C. (2003) Arch. Biochem. Biophys. 414, 130-140. - PubMed
-
- Parry, M. A. J., Andralojc, R. A. C., Mitchell, P. J., Madgwick, P. J. & Keys, A. J. (2003) J. Exp. Bot. 54, 1321-1333. - PubMed
-
- Chen, Z. & Spreitzer, R. J. (1992) Photosynth. Res. 31, 157-164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

