Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides
- PMID: 16282466
- PMCID: PMC1287559
- DOI: 10.1128/JVI.79.23.14660-14667.2005
Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides
Abstract
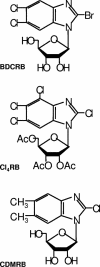
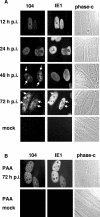
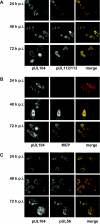
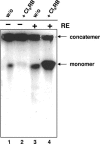
Herpesvirus DNA replication leads to unit length genomes that are translocated into preformed procapsids through a unique portal vertex. The translocation is performed by the terminase that cleaves the DNA and powers the insertion by its ATPase activity. Recently, we demonstrated that the putative human cytomegalovirus (HCMV) portal protein, pUL104, also forms high-molecular-weight complexes. Analyses now have been performed to determine the intracellular localization and identification of interaction partners of pUL104. In infected cells, HCMV pUL104 was found to be predominantly localized throughout the nucleus as well as in cytoplasmic clusters at late times of infection. The latter localization was abolished by phosphonoacetic acid, an inhibitor of viral DNA replication. Immunofluorescence revealed that pUL104 colocalized with pUL56, the large subunit of the HCMV terminase. Specific association of in vitro translated pUL104 with the carboxy-terminal half of GST-UL56C was detected. By using coimmunoprecipitations a direct interaction with pUL56 was confirmed. In addition, this interaction was no longer detected when the benzimidazole-D-nucleosides BDCRB or Cl4RB were added, thus indicating that these HCMV inhibitors block the insertion of the DNA into the capsid by preventing a necessary interaction of pUL56 with the portal. Electron microscopy revealed that in the presence of Cl4RB DNA is not packaged into capsids and these capsids failed to egress from the nucleus. Furthermore, pulsed-field gel electrophoresis showed that DNA concatemers synthesized in the presence of the compound failed to be processed.
Figures



Similar articles
-
Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity.Antivir Ther. 2008;13(5):643-54. Antivir Ther. 2008. PMID: 18771048
-
Tetrahalogenated benzimidazole D-ribonucleosides are active against rat cytomegalovirus.Antiviral Res. 2017 Jan;137:102-107. doi: 10.1016/j.antiviral.2016.11.012. Epub 2016 Nov 18. Antiviral Res. 2017. PMID: 27871886
-
Targeting the terminase: An important step forward in the treatment and prophylaxis of human cytomegalovirus infections.Antiviral Res. 2019 Jan;161:116-124. doi: 10.1016/j.antiviral.2018.11.005. Epub 2018 Nov 23. Antiviral Res. 2019. PMID: 30472161 Review.
-
Identification of the interaction domain of the small terminase subunit pUL89 with the large subunit pUL56 of human cytomegalovirus.Biochemistry. 2006 Jul 25;45(29):8855-63. doi: 10.1021/bi0600796. Biochemistry. 2006. PMID: 16846228
-
Clinical development of letermovir and maribavir: Overview of human cytomegalovirus drug resistance.Antiviral Res. 2019 Mar;163:91-105. doi: 10.1016/j.antiviral.2019.01.011. Epub 2019 Jan 25. Antiviral Res. 2019. PMID: 30690043 Review.
Cited by
-
Direct visualization of the putative portal in the Kaposi's sarcoma-associated herpesvirus capsid by cryoelectron tomography.J Virol. 2007 Apr;81(7):3640-4. doi: 10.1128/JVI.02254-06. Epub 2007 Jan 10. J Virol. 2007. PMID: 17215290 Free PMC article.
-
Computational modeling of protracted HCMV replication using genome substrates and protein temporal profiles.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2201787119. doi: 10.1073/pnas.2201787119. Epub 2022 Aug 22. Proc Natl Acad Sci U S A. 2022. PMID: 35994667 Free PMC article.
-
Intracellular Distribution of Capsid-Associated pUL77 of Human Cytomegalovirus and Interactions with Packaging Proteins and pUL93.J Virol. 2016 Jun 10;90(13):5876-5885. doi: 10.1128/JVI.00351-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27053556 Free PMC article.
-
Viral and host control of cytomegalovirus maturation.Trends Microbiol. 2012 Aug;20(8):392-401. doi: 10.1016/j.tim.2012.04.008. Epub 2012 May 23. Trends Microbiol. 2012. PMID: 22633075 Free PMC article. Review.
-
Susceptibilities of human cytomegalovirus clinical isolates and other herpesviruses to new acetylated, tetrahalogenated benzimidazole D-ribonucleosides.Antimicrob Agents Chemother. 2009 Dec;53(12):5095-101. doi: 10.1128/AAC.00809-09. Epub 2009 Sep 28. Antimicrob Agents Chemother. 2009. PMID: 19786605 Free PMC article.
References
-
- Anders, D. G, and L. A. McCue. 1996. The human cytomegalovirus genes and proteins required for DNA synthesis. Intervirology 39:378-388. - PubMed
-
- Andreoni, M., M. Faircloth, L. Vulger, and, W. J., Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. - PubMed
-
- Bazinet, C., J. Benbasat, J. King, J. M. Carazo, and J. L. Carrascosa. 1988. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry 27:1849-1856. - PubMed
-
- Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Mircrobiol. 43:267-292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

