Human parainfluenza virus type 4 is incapable of evading the interferon-induced antiviral effect
- PMID: 16282476
- PMCID: PMC1287573
- DOI: 10.1128/JVI.79.23.14756-14768.2005
Human parainfluenza virus type 4 is incapable of evading the interferon-induced antiviral effect
Abstract
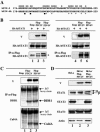
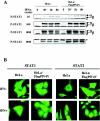
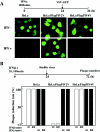
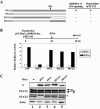
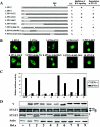
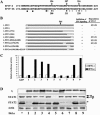
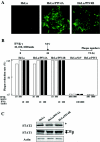
The V proteins of some paramyxoviruses have developed the ability to efficiently inactivate STAT protein function as a countermeasure for evading interferon (IFN) responses. Human parainfluenza virus type 4 (hPIV4) is one of the rubulaviruses, which are members of the family Paramyxoviridae, and has a V protein with a highly conserved cysteine-rich domain that is the hallmark of paramyxovirus V proteins. In order to study the function of the hPIV4 V protein, we established HeLa cells expressing the hPIV4A V protein (HeLa/FlagPIV4V). The hPIV4 V protein had no ability to reduce the level of STAT1 or STAT2, although it associated with STAT1, STAT2, DDB1, and Cul4A. It interfered with neither STAT1 and STAT2 tyrosine phosphorylation nor IFN-induced STAT nuclear accumulation. In addition, HeLa/FlagPIV4V cells are fully sensitive to both beta interferon (IFN-beta) and IFN-gamma, indicating that the hPIV4 V protein has no ability to block IFN-induced signaling. We further established HeLa cells expressing various chimeric proteins between the hPIV2 and hPIV4A V proteins. The lack of IFN-antagonistic activity of the hPIV4 V protein is caused by both the P/V common and V-specific domains. At least two regions (amino acids [aa] 32 to 45 and aa 143 to 164) of hPIV4 V in the P/V common domain and one region (aa 200 to 212) of the C terminus are involved in the inability to evade the IFN-induced signaling. Moreover, we established HeLa cells persistently infected with hPIV4 to make sure of the inability to escape IFN and confirmed that hPIV4 is the only paramyxovirus analyzed to date that can't evade the IFN-induced antiviral responses.
Figures
Similar articles
-
High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons.J Virol. 2001 Oct;75(19):9165-76. doi: 10.1128/JVI.75.19.9165-9176.2001. J Virol. 2001. PMID: 11533180 Free PMC article.
-
Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response.Virology. 2000 Apr 10;269(2):383-90. doi: 10.1006/viro.2000.0240. Virology. 2000. PMID: 10753717
-
Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation.J Virol. 2002 Nov;76(22):11476-83. doi: 10.1128/jvi.76.22.11476-11483.2002. J Virol. 2002. PMID: 12388709 Free PMC article.
-
Paramyxovirus strategies for evading the interferon response.Rev Med Virol. 2002 Nov-Dec;12(6):337-57. doi: 10.1002/rmv.357. Rev Med Virol. 2002. PMID: 12410527 Review.
-
Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein.Eur J Biochem. 2004 Dec;271(23-24):4621-8. doi: 10.1111/j.1432-1033.2004.04425.x. Eur J Biochem. 2004. PMID: 15606749 Review.
Cited by
-
The V protein of Tioman virus is incapable of blocking type I interferon signaling in human cells.PLoS One. 2013;8(1):e53881. doi: 10.1371/journal.pone.0053881. Epub 2013 Jan 14. PLoS One. 2013. PMID: 23342031 Free PMC article.
-
Paramyxovirus disruption of interferon signal transduction: STATus report.J Interferon Cytokine Res. 2009 Sep;29(9):531-7. doi: 10.1089/jir.2009.0070. J Interferon Cytokine Res. 2009. PMID: 19694544 Free PMC article. Review.
-
IFN-gamma regulation of ICAM-1 receptors in bronchial epithelial cells: soluble ICAM-1 release inhibits human rhinovirus infection.J Inflamm (Lond). 2008 Jun 5;5:8. doi: 10.1186/1476-9255-5-8. J Inflamm (Lond). 2008. PMID: 18534017 Free PMC article.
-
Antagonism of innate immunity by paramyxovirus accessory proteins.Viruses. 2009 Dec;1(3):574-593. doi: 10.3390/v1030574. Epub 2009 Oct 28. Viruses. 2009. PMID: 21994561 Free PMC article.
-
Type I and Type II Interferon Antagonism Strategies Used by Paramyxoviridae: Previous and New Discoveries, in Comparison.Viruses. 2022 May 21;14(5):1107. doi: 10.3390/v14051107. Viruses. 2022. PMID: 35632848 Free PMC article. Review.
References
-
- Bando, H., K. Kondo, M. Kawano, H. Komada, M. Tsurudome, M. Nishio, and Y. Ito. 1990. Molecular cloning and sequence analysis of human parainfluenza type 4A virus HN gene: its irregularities on structure and activities. Virology 175:307-312. - PubMed
-
- Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

