Commensurate distances and similar motifs in genetic congruence and protein interaction networks in yeast
- PMID: 16283923
- PMCID: PMC1310633
- DOI: 10.1186/1471-2105-6-270
Commensurate distances and similar motifs in genetic congruence and protein interaction networks in yeast
Abstract
Background: In a genetic interaction, the phenotype of a double mutant differs from the combined phenotypes of the underlying single mutants. When the single mutants have no growth defect, but the double mutant is lethal or exhibits slow growth, the interaction is termed synthetic lethality or synthetic fitness. These genetic interactions reveal gene redundancy and compensating pathways. Recently available large-scale data sets of genetic interactions and protein interactions in Saccharomyces cerevisiae provide a unique opportunity to elucidate the topological structure of biological pathways and how genes function in these pathways.
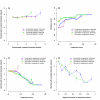
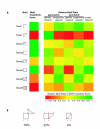
Results: We have defined congruent genes as pairs of genes with similar sets of genetic interaction partners and constructed a genetic congruence network by linking congruent genes. By comparing path lengths in three types of networks (genetic interaction, genetic congruence, and protein interaction), we discovered that high genetic congruence not only exhibits correlation with direct protein interaction linkage but also exhibits commensurate distance with the protein interaction network. However, consistent distances were not observed between genetic and protein interaction networks. We also demonstrated that congruence and protein networks are enriched with motifs that indicate network transitivity, while the genetic network has both transitive (triangle) and intransitive (square) types of motifs. These results suggest that robustness of yeast cells to gene deletions is due in part to two complementary pathways (square motif) or three complementary pathways, any two of which are required for viability (triangle motif).
Conclusion: Genetic congruence is superior to genetic interaction in prediction of protein interactions and function associations. Genetically interacting pairs usually belong to parallel compensatory pathways, which can generate transitive motifs (any two of three pathways needed) or intransitive motifs (either of two pathways needed).
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

