Structure of a V3-containing HIV-1 gp120 core
- PMID: 16284180
- PMCID: PMC2408531
- DOI: 10.1126/science.1118398
Structure of a V3-containing HIV-1 gp120 core
Abstract
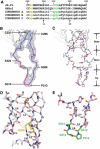
The third variable region (V3) of the HIV-1 gp120 envelope glycoprotein is immunodominant and contains features essential for coreceptor binding. We determined the structure of V3 in the context of an HIV-1 gp120 core complexed to the CD4 receptor and to the X5 antibody at 3.5 angstrom resolution. Binding of gp120 to cell-surface CD4 would position V3 so that its coreceptor-binding tip protrudes 30 angstroms from the core toward the target cell membrane. The extended nature and antibody accessibility of V3 explain its immunodominance. Together, the results provide a structural rationale for the role of V3 in HIV entry and neutralization.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

