The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis
- PMID: 16284305
- PMCID: PMC1315373
- DOI: 10.1105/tpc.105.036814
The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis
Abstract
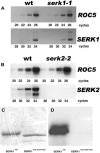
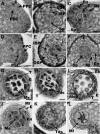
The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) family of plasma membrane receptors consists of five closely related members. The SERK1 and SERK2 genes show a complex expression pattern throughout development. Both are expressed in anther primordia up to the second parietal division. After this point, expression ceases in the sporocytes and is continued in the tapetum and middle layer precursors. Single knockout mutants of SERK1 and SERK2 show no obvious phenotypes. Double mutants of SERK1 and SERK2 are completely male sterile due to a failure in tapetum specification. Fertility can be restored by a single copy of either gene. The SERK1 and SERK2 proteins can form homodimers or heterodimers in vivo, suggesting they are interchangeable in the SERK1/SERK2 signaling complex.
Figures






References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Becker, W., Bergmann, A., Hink, M.A., Konig, K., Benndorf, K., and Biscup, C. (2004). Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc. Res. Tech. 63, 58–66. - PubMed
-
- Borst, J.W., Hink, M., van Hoek, A., and Visser, A.J.W.G. (2003). Multiphoton microspectroscopy in living plant cells. In Multiphoton Microscopy in the Biomedical Sciences III, A. Periasamy and P.T. So, eds (Bellingham, WA: SPIE), pp. 231–238.
-
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12, 1718–1727. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

