Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis
- PMID: 16284311
- PMCID: PMC1315367
- DOI: 10.1105/tpc.105.035840
Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis
Abstract
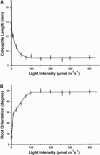
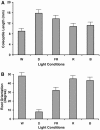
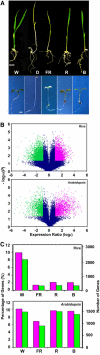
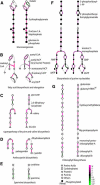
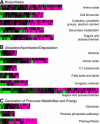
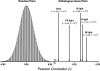
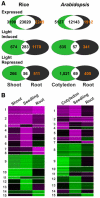
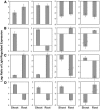
Genome-wide 70-mer oligonucleotide microarrays of rice (Oryza sativa) and Arabidopsis thaliana were used to profile genome expression changes during light-regulated seedling development. We estimate that the expression of approximately 20% of the genome in both rice and Arabidopsis seedlings is regulated by white light. Qualitatively similar expression profiles from seedlings grown under different light qualities were observed in both species; however, a quantitatively weaker effect on genome expression was observed in rice. Most metabolic pathways exhibited qualitatively similar light regulation in both species with a few species-specific differences. Global comparison of expression profiles between rice and Arabidopsis reciprocal best-matched gene pairs revealed a higher correlation of genome expression patterns in constant light than in darkness, suggesting that the genome expression profile of photomorphogenesis is more conserved. Transcription factor gene expression under constant light exposure was poorly conserved between the two species, implying a faster-evolving rate of transcription factor gene expression in light-grown plants. Organ-specific expression profiles during seedling photomorphogenesis provide genome-level evidence for divergent light effects in different higher plant organs. Finally, overrepresentation of specific promoter motifs in root- and leaf-specific light-regulated genes in both species suggests that these cis-elements are important for gene expression responses to light.
Figures



Similar articles
-
Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation.Plant Physiol. 2011 Mar;155(3):1353-66. doi: 10.1104/pp.110.166801. Epub 2011 Jan 10. Plant Physiol. 2011. PMID: 21220763 Free PMC article.
-
Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis.Plant Cell. 2002 Oct;14(10):2383-98. doi: 10.1105/tpc.004416. Plant Cell. 2002. PMID: 12368493 Free PMC article.
-
Rapid, organ-specific transcriptional responses to light regulate photomorphogenic development in dicot seedlings.Plant Physiol. 2011 Aug;156(4):2124-40. doi: 10.1104/pp.111.179416. Epub 2011 Jun 7. Plant Physiol. 2011. PMID: 21653191 Free PMC article.
-
Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species.Plant Physiol. 2004 Jun;135(2):622-9. doi: 10.1104/pp.104.040170. Plant Physiol. 2004. PMID: 15208410 Free PMC article. Review.
-
Toward an understanding of the detection and function of R-loops in plants.J Exp Bot. 2021 Sep 30;72(18):6110-6122. doi: 10.1093/jxb/erab280. J Exp Bot. 2021. PMID: 34115858 Review.
Cited by
-
The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels.Hortic Res. 2021 Apr 1;8(1):83. doi: 10.1038/s41438-021-00523-0. Hortic Res. 2021. PMID: 33790264 Free PMC article.
-
Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis.Plant Cell. 2009 Dec;21(12):3732-48. doi: 10.1105/tpc.109.066845. Epub 2009 Dec 11. Plant Cell. 2009. PMID: 20008096 Free PMC article.
-
BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21.Plant Physiol. 2011 Aug;156(4):2109-23. doi: 10.1104/pp.111.177139. Epub 2011 Jun 1. Plant Physiol. 2011. PMID: 21632973 Free PMC article.
-
Plant MetGenMAP: an integrative analysis system for plant systems biology.Plant Physiol. 2009 Dec;151(4):1758-68. doi: 10.1104/pp.109.145169. Epub 2009 Oct 9. Plant Physiol. 2009. PMID: 19819981 Free PMC article.
-
Sequence and expression analysis of the thioredoxin protein gene family in rice.Mol Genet Genomics. 2008 Aug;280(2):139-51. doi: 10.1007/s00438-008-0351-4. Epub 2008 May 20. Mol Genet Genomics. 2008. PMID: 18491141
References
-
- Basu, D., Dehesh, K., Schneider-Poetsch, H.J., Harrington, S.E., McCouch, S.R., and Quail, P.H. (2000). Rice PHYC gene: Structure, expression, map position and evolution. Plant Mol. Biol. 44, 27–42. - PubMed
-
- Blecken, J., Weisshaar, B., and Herzfeld, F. (1994). Two distinct cis-acting elements are involved in light-dependent activation of the pea elip promoter. Mol. Gen. Genet. 245, 371–379. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

