NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii
- PMID: 16284312
- PMCID: PMC1315378
- DOI: 10.1105/tpc.105.035774
NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii
Abstract
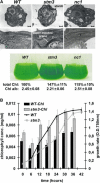
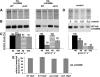
Photosynthetic organisms respond to changes in ambient light by modulating the size and composition of their light-harvesting complexes, which in the case of the green alga Chlamydomonas reinhardtii consists of >15 members of a large extended family of chlorophyll binding subunits. How their expression is coordinated is unclear. Here, we describe the analysis of an insertion mutant, state transitions mutant3 (stm3), which we show has increased levels of LHCBM subunits associated with the light-harvesting antenna of photosystem II. The mutated nuclear gene in stm3 encodes the RNA binding protein NAB1 (for putative nucleic acid binding protein). In vitro and in vivo RNA binding and protein expression studies have confirmed that NAB1 differentially binds to LHCBM mRNA in a subpolysomal high molecular weight RNA-protein complex. Binding of NAB1 stabilizes LHCBM mRNA at the preinitiation level via sequestration and thereby represses translation. The specificity and affinity of binding are determined by an RNA sequence motif similar to that used by the Xenopus laevis translation repressor FRGY2, which is conserved to varying degrees in the LHCBM gene family. We conclude from our results that NAB1 plays an important role in controlling the expression of the light-harvesting antenna of photosystem II at the posttranscriptional level. The similarity of NAB1 and FRGY2 of Xenopus implies the existence of similar RNA-masking systems in animals and plants.
Figures





Similar articles
-
Integration of carbon assimilation modes with photosynthetic light capture in the green alga Chlamydomonas reinhardtii.Mol Plant. 2014 Oct;7(10):1545-59. doi: 10.1093/mp/ssu083. Epub 2014 Jul 18. Mol Plant. 2014. PMID: 25038233
-
Cysteine modification of a specific repressor protein controls the translational status of nucleus-encoded LHCII mRNAs in Chlamydomonas.Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13290-5. doi: 10.1073/pnas.0900670106. Epub 2009 Aug 3. Proc Natl Acad Sci U S A. 2009. PMID: 19666611 Free PMC article.
-
Solution structure of the RNA-binding cold-shock domain of the Chlamydomonas reinhardtii NAB1 protein and insights into RNA recognition.Biochem J. 2015 Jul 1;469(1):97-106. doi: 10.1042/BJ20150217. Epub 2015 Apr 28. Biochem J. 2015. PMID: 25919092
-
A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii.Curr Genet. 2004 Feb;45(2):61-75. doi: 10.1007/s00294-003-0460-x. Epub 2003 Dec 2. Curr Genet. 2004. PMID: 14652691 Review.
-
Structural analysis and comparison of light-harvesting complexes I and II.Biochim Biophys Acta Bioenerg. 2020 Apr 1;1861(4):148038. doi: 10.1016/j.bbabio.2019.06.010. Epub 2019 Jun 20. Biochim Biophys Acta Bioenerg. 2020. PMID: 31229568 Review.
Cited by
-
Cell Surface and Membrane Engineering: Emerging Technologies and Applications.J Funct Biomater. 2015 Jun 18;6(2):454-85. doi: 10.3390/jfb6020454. J Funct Biomater. 2015. PMID: 26096148 Free PMC article. Review.
-
State transitions revisited-a buffering system for dynamic low light acclimation of Arabidopsis.Plant Mol Biol. 2006 Nov;62(4-5):779-93. doi: 10.1007/s11103-006-9044-8. Epub 2006 Aug 1. Plant Mol Biol. 2006. PMID: 16897465
-
Carbon Supply and Photoacclimation Cross Talk in the Green Alga Chlamydomonas reinhardtii.Plant Physiol. 2016 Nov;172(3):1494-1505. doi: 10.1104/pp.16.01310. Epub 2016 Sep 16. Plant Physiol. 2016. PMID: 27637747 Free PMC article.
-
Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals.J Exp Bot. 2012 Feb;63(3):1297-313. doi: 10.1093/jxb/err356. Epub 2011 Nov 29. J Exp Bot. 2012. PMID: 22131159 Free PMC article.
-
CO2 supply modulates lipid remodelling, photosynthetic and respiratory activities in Chlorella species.Plant Cell Environ. 2021 Sep;44(9):2987-3001. doi: 10.1111/pce.14074. Epub 2021 May 17. Plant Cell Environ. 2021. PMID: 33931891 Free PMC article.
References
-
- Allen, J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098, 275–335. - PubMed
-
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6, 317–326. - PubMed
-
- Allen, K.D., and Staehelin, L.A. (1991). Resolution of 16 to 20 chlorophyll protein complexes using a low ionic-strength native green gel system. Anal. Biochem. 194, 214–222. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

