AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis
- PMID: 16284313
- PMCID: PMC1315382
- DOI: 10.1105/tpc.105.035659
AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis
Abstract
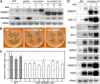
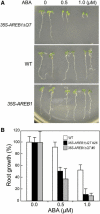
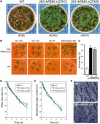
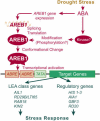
ABSCISIC ACID-RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1) (i.e., ABF2) is a basic domain/leucine zipper transcription factor that binds to the abscisic acid (ABA)-responsive element (ABRE) motif in the promoter region of ABA-inducible genes. Here, we show that expression of the intact AREB1 gene on its own is insufficient to lead to expression of downstream genes under normal growth conditions. To overcome the masked transactivation activity of AREB1, we created an activated form of AREB1 (AREB1DeltaQT). AREB1DeltaQT-overexpressing plants showed ABA hypersensitivity and enhanced drought tolerance, and eight genes with two or more ABRE motifs in the promoter regions in two groups were greatly upregulated: late embryogenesis abundant class genes and ABA- and drought stress-inducible regulatory genes. By contrast, an areb1 null mutant and a dominant loss-of-function mutant of AREB1 (AREB1:RD) with a repression domain exhibited ABA insensitivity. Furthermore, AREB1:RD plants displayed reduced survival under dehydration, and three of the eight greatly upregulated genes were downregulated, including genes for linker histone H1 and AAA ATPase, which govern gene expression and multiple cellular activities through protein folding, respectively. Thus, these data suggest that AREB1 regulates novel ABRE-dependent ABA signaling that enhances drought tolerance in vegetative tissues.
Figures



References
-
- Ascenzi, R., and Gantt, J.S. (1997). A drought-inducible histone gene in Arabidopsis is a member of a distinct class of plant linker histone variants. Plant Mol. Biol. 34 629–641. - PubMed
-
- Ascenzi, R., and Gantt, J.S. (1999). Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana. Plant Mol. Biol. 41 159–169. - PubMed
-
- Bray, E.A. (1994). Alterations in gene expression in response to water deficit. In Stress-Induced Gene Expression in Plants, A.S. Basra, ed (Amsterdam: Harwood Academic), pp. 1–23.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

