Direct observation of base-pair stepping by RNA polymerase
- PMID: 16284617
- PMCID: PMC1356566
- DOI: 10.1038/nature04268
Direct observation of base-pair stepping by RNA polymerase
Abstract
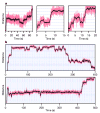
During transcription, RNA polymerase (RNAP) moves processively along a DNA template, creating a complementary RNA. Here we present the development of an ultra-stable optical trapping system with ångström-level resolution, which we used to monitor transcriptional elongation by single molecules of Escherichia coli RNAP. Records showed discrete steps averaging 3.7 +/- 0.6 A, a distance equivalent to the mean rise per base found in B-DNA. By combining our results with quantitative gel analysis, we conclude that RNAP advances along DNA by a single base pair per nucleotide addition to the nascent RNA. We also determined the force-velocity relationship for transcription at both saturating and sub-saturating nucleotide concentrations; fits to these data returned a characteristic distance parameter equivalent to one base pair. Global fits were inconsistent with a model for movement incorporating a power stroke tightly coupled to pyrophosphate release, but consistent with a brownian ratchet model incorporating a secondary NTP binding site.
Figures




References
-
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. - PubMed
-
- Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. - PubMed
-
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. - PubMed
-
- Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

