p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death
- PMID: 16286508
- PMCID: PMC2171557
- DOI: 10.1083/jcb.200507002
p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death
Abstract
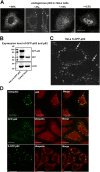
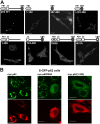
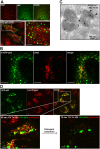
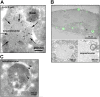
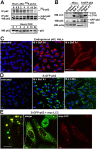
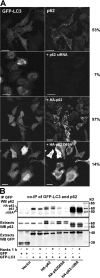
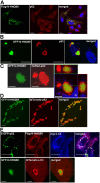
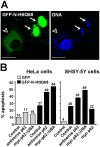
Autophagic degradation of ubiquitinated protein aggregates is important for cell survival, but it is not known how the autophagic machinery recognizes such aggregates. In this study, we report that polymerization of the polyubiquitin-binding protein p62/SQSTM1 yields protein bodies that either reside free in the cytosol and nucleus or occur within autophagosomes and lysosomal structures. Inhibition of autophagy led to an increase in the size and number of p62 bodies and p62 protein levels. The autophagic marker light chain 3 (LC3) colocalized with p62 bodies and co-immunoprecipitated with p62, suggesting that these two proteins participate in the same complexes. The depletion of p62 inhibited recruitment of LC3 to autophagosomes under starvation conditions. Strikingly, p62 and LC3 formed a shell surrounding aggregates of mutant huntingtin. Reduction of p62 protein levels or interference with p62 function significantly increased cell death that was induced by the expression of mutant huntingtin. We suggest that p62 may, via LC3, be involved in linking polyubiquitinated protein aggregates to the autophagy machinery.
Figures
References
-
- Arrasate, M., S. Mitra, E.S. Schweitzer, M.R. Segal, and S. Finkbeiner. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810. - PubMed
-
- Bence, N.F., R.M. Sampat, and R.R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

