Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells
- PMID: 16286661
- PMCID: PMC1283154
- DOI: 10.1073/pnas.0502129102
Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells
Abstract
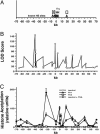
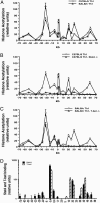
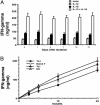
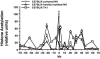
Local histone acetylation of promoters precedes transcription of many genes. Extended histone hyperacetylation at great distances from coding regions of genes also occurs during active transcription of gene families or individual genes and may reflect developmental processes that mark genes destined for cell-specific transcription, nuclear signaling processes that are required for active transcription, or both. To distinguish between these, we compared long-range histone acetylation patterns across the Ifng gene region in natural killer (NK) cells and T cells that were or were not actively transcribing the Ifng gene. In T cells, long-range histone acetylation depended on stimulation that drives both T helper (Th) 1 differentiation and active transcription, and it depended completely or partially on the presence of Stat4 or T-bet, respectively, two transcription factors that are required for Th1 lineage commitment. In contrast, in the absence of stimulation and active transcription, similar histone hyperacetylated domains were found in NK cells. Additional proximal domains were hyperacetylated after stimulation of transcription. We hypothesize that formation of extended histone hyperacetylated domains across the Ifng gene region represents a developmental mechanism that marks this gene for cell- or stimulus-specific transcription.
Figures
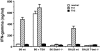

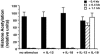
References
-
- Brown, K. E., Amoils, S., Horn, J. M., Buckle, V. J., Higgs, D. R., Merkenschlager, M. & Fisher, A. G. (2001) Nat. Cell Biol. 3, 602-606. - PubMed
-
- Ansel, K. M., Lee, D. U. & Rao, A. (2003) Nat. Immunol. 4, 616-623. - PubMed
-
- Mossman, T. R. & Coffman, R. L. (1989) Ann. Rev. Immunol. 7, 145-173. - PubMed
-
- Seder, R. A. & Paul, W. E. (1994) Ann. Rev. Immunol. 12, 635-673. - PubMed
-
- Glimcher, L. H. & Murphy, K. (2000) Genes Dev. 14, 1693-1711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

