Loss of SOD1 and LYS7 sensitizes Saccharomyces cerevisiae to hydroxyurea and DNA damage agents and downregulates MEC1 pathway effectors
- PMID: 16287844
- PMCID: PMC1291217
- DOI: 10.1128/MCB.25.23.10273-10285.2005
Loss of SOD1 and LYS7 sensitizes Saccharomyces cerevisiae to hydroxyurea and DNA damage agents and downregulates MEC1 pathway effectors
Abstract
Aerobic metabolism produces reactive oxygen species, including superoxide anions, which cause DNA damage unless removed by scavengers such as superoxide dismutases. We show that loss of the Cu,Zn-dependent superoxide dismutase, SOD1, or its copper chaperone, LYS7, confers oxygen-dependent sensitivity to replication arrest and DNA damage in Saccharomyces cerevisiae. We also find that sod1Delta strains, and to a lesser extent lys7Delta strains, when arrested with hydroxyurea (HU) show reduced induction of the MEC1 pathway effector Rnr3p and of Hug1p. The HU sensitivity of sod1Delta and lys7Delta strains is suppressed by overexpression of TKL1, a transketolase that generates NADPH, which balances redox in the cell and is required for ribonucleotide reductase activity. Our results suggest that the MEC1 pathway in sod1Delta mutant strains is sensitive to the altered cellular redox state due to increased superoxide anions and establish a new relationship between SOD1, LYS7, and the MEC1-mediated checkpoint response to replication arrest and DNA damage in S. cerevisiae.
Figures


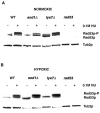
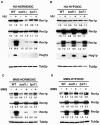




Similar articles
-
Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway.Mol Cell Biol. 2004 Nov;24(22):10016-25. doi: 10.1128/MCB.24.22.10016-10025.2004. Mol Cell Biol. 2004. PMID: 15509802 Free PMC article.
-
The yeast copper response is regulated by DNA damage.Mol Cell Biol. 2013 Oct;33(20):4041-50. doi: 10.1128/MCB.00116-13. Epub 2013 Aug 19. Mol Cell Biol. 2013. PMID: 23959798 Free PMC article.
-
Lack of superoxide dismutase in a rad51 mutant exacerbates genomic instability and oxidative stress-mediated cytotoxicity in Saccharomyces cerevisiae.Free Radic Biol Med. 2018 Dec;129:97-106. doi: 10.1016/j.freeradbiomed.2018.09.015. Epub 2018 Sep 14. Free Radic Biol Med. 2018. PMID: 30223018
-
A copper connection to the uptake of platinum anticancer drugs.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):13963-5. doi: 10.1073/pnas.232574299. Epub 2002 Oct 21. Proc Natl Acad Sci U S A. 2002. PMID: 12391309 Free PMC article. Review. No abstract available.
-
Coping with and recovering from hydroxyurea-induced replication fork arrest in budding yeast.Cold Spring Harb Symp Quant Biol. 2000;65:333-42. doi: 10.1101/sqb.2000.65.333. Cold Spring Harb Symp Quant Biol. 2000. PMID: 12760047 Review. No abstract available.
Cited by
-
Synthetic lethal interaction between oxidative stress response and DNA damage repair in the budding yeast and its application to targeted anticancer therapy.J Microbiol. 2019 Jan;57(1):9-17. doi: 10.1007/s12275-019-8475-2. Epub 2018 Dec 29. J Microbiol. 2019. PMID: 30594981 Review.
-
Nuclear SOD1 in Growth Control, Oxidative Stress Response, Amyotrophic Lateral Sclerosis, and Cancer.Antioxidants (Basel). 2022 Feb 21;11(2):427. doi: 10.3390/antiox11020427. Antioxidants (Basel). 2022. PMID: 35204309 Free PMC article. Review.
-
Excess copper induces anoxygenic photosynthesis in Anabaena doliolum: a homology based proteomic assessment of its survival strategy.Photosynth Res. 2008 Apr;96(1):61-74. doi: 10.1007/s11120-007-9285-7. Epub 2007 Dec 29. Photosynth Res. 2008. PMID: 18165907
-
The Cell Killing Mechanisms of Hydroxyurea.Genes (Basel). 2016 Nov 17;7(11):99. doi: 10.3390/genes7110099. Genes (Basel). 2016. PMID: 27869662 Free PMC article. Review.
-
Heme deficiency sensitizes yeast cells to oxidative stress induced by hydroxyurea.J Biol Chem. 2017 Jun 2;292(22):9088-9103. doi: 10.1074/jbc.M117.781211. Epub 2017 Apr 4. J Biol Chem. 2017. PMID: 28377506 Free PMC article.
References
-
- Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
-
- Aruoma, O. I., B. Halliwell, B. M. Hoey, and J. Butler. 1989. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic. Biol. Med. 6:593-597. - PubMed
-
- Assfalg, M., L. Banci, I. Bertini, P. Turano, and P. R. Vasos. 2003. Superoxide dismutase folding/unfolding pathway: role of the metal ions in modulating structural and dynamical features. J. Mol. Biol. 330:145-158. - PubMed
-
- Barzilai, A., G. Rotman, and Y. Shiloh. 2002. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair 1:3-25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
