The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14
- PMID: 16287850
- PMCID: PMC1291222
- DOI: 10.1128/MCB.25.23.10352-10364.2005
The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14
Abstract
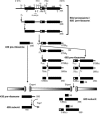
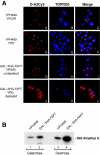
One of the proteins identified as being involved in ribosome biogenesis by high-throughput studies, a putative P-loop-type kinase termed Fap7 (YDL166c), was shown to be required for the conversion of 20S pre-rRNA to 18S rRNA. However, the mechanism underlying this function has remained unclear. Here we demonstrate that Fap7 is strictly required for cleavage of the 20S pre-rRNA at site D in the cytoplasm. Genetic depletion of Fap7 causes accumulation of only the 20S pre-rRNA, which could be detected not only in 43S preribosomes but also in 80S-sized complexes. Fap7 is not a structural component of 43S preribosomes but likely transiently interacts with them by directly binding to Rps14, a ribosomal protein that is found near the 3' end of the 18S rRNA. Consistent with an NTPase activity, conserved residues predicted to be required for nucleoside triphosphate (NTP) hydrolysis are essential for Fap7 function in vivo. We propose that Fap7 mediates cleavage of the 20S pre-rRNA at site D by directly interacting with Rps14 and speculate that it is an enzyme that functions as an NTP-dependent molecular switch in 18S rRNA maturation.
Figures





References
-
- Antunez de Mayolo, P., and J. L. Woolford, Jr. 2003. Interactions of yeast ribosomal protein rpS14 with RNA. J. Mol. Biol. 333:697-709. - PubMed
-
- Arcus, V. L., K. Backbro, A. Roos, E. L. Daniel, and E. N. Baker. 2004. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 279:16471-16478. - PubMed
-
- Brand, R. C., J. Klootwijk, T. J. Van Steenbergen, A. J. De Kok, and R. J. Planta. 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75:311-318. - PubMed
-
- Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
