Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors
- PMID: 16287856
- PMCID: PMC1291231
- DOI: 10.1128/MCB.25.23.10433-10441.2005
Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors
Abstract
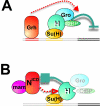
Notch signal transduction centers on a conserved DNA-binding protein called Suppressor of Hairless [Su(H)] in Drosophila species. In the absence of Notch activation, target genes are repressed by Su(H) acting in conjunction with a partner, Hairless, which contains binding motifs for two global corepressors, CtBP and Groucho (Gro). Usually these corepressors are thought to act via different mechanisms; complexed with other transcriptional regulators, they function independently and/or redundantly. Here we have investigated the requirement for Gro and CtBP in Hairless-mediated repression. Unexpectedly, we find that mutations inactivating one or the other binding motif can have detrimental effects on Hairless similar to those of mutations that inactivate both motifs. These results argue that recruitment of one or the other corepressor is not sufficient to confer repression in the context of the Hairless-Su(H) complex; Gro and CtBP need to function in combination. In addition, we demonstrate that Hairless has a second mode of repression that antagonizes Notch intracellular domain and is independent of Gro or CtBP binding.
Figures

Similar articles
-
Fine tuning of Notch signaling by differential co-repressor recruitment during eye development of Drosophila.Hereditas. 2011 Jun;148(3):77-84. doi: 10.1111/j.1601-5223.2011.02221.x. Epub 2011 May 26. Hereditas. 2011. PMID: 21756252
-
Molecular analysis of the notch repressor-complex in Drosophila: characterization of potential hairless binding sites on suppressor of hairless.PLoS One. 2011;6(11):e27986. doi: 10.1371/journal.pone.0027986. Epub 2011 Nov 18. PLoS One. 2011. PMID: 22125648 Free PMC article.
-
Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless.Genes Dev. 2002 Aug 1;16(15):1964-76. doi: 10.1101/gad.987402. Genes Dev. 2002. PMID: 12154126 Free PMC article.
-
Hairless: the ignored antagonist of the Notch signalling pathway.Hereditas. 2006 Dec;143(2006):212-21. doi: 10.1111/j.2007.0018-0661.01971.x. Hereditas. 2006. PMID: 17362357 Review.
-
Notch pathway: making sense of suppressor of hairless.Curr Biol. 2001 Mar 20;11(6):R217-21. doi: 10.1016/s0960-9822(01)00109-9. Curr Biol. 2001. PMID: 11301266 Review.
Cited by
-
In vivo analysis of internal ribosome entry at the Hairless locus by genome engineering in Drosophila.Sci Rep. 2016 Oct 7;6:34881. doi: 10.1038/srep34881. Sci Rep. 2016. PMID: 27713501 Free PMC article.
-
Novel Genome-Engineered H Alleles Differentially Affect Lateral Inhibition and Cell Dichotomy Processes during Bristle Organ Development.Genes (Basel). 2024 Apr 26;15(5):552. doi: 10.3390/genes15050552. Genes (Basel). 2024. PMID: 38790181 Free PMC article.
-
Three distinct mechanisms, Notch instructive, permissive, and independent, regulate the expression of two different pericardial genes to specify cardiac cell subtypes.PLoS One. 2020 Oct 27;15(10):e0241191. doi: 10.1371/journal.pone.0241191. eCollection 2020. PLoS One. 2020. PMID: 33108408 Free PMC article.
-
Planar cell polarity controls directional Notch signaling in the Drosophila leg.Development. 2012 Jul;139(14):2584-93. doi: 10.1242/dev.077446. Development. 2012. PMID: 22736244 Free PMC article.
-
Notch signaling: switching an oncogene to a tumor suppressor.Blood. 2014 Apr 17;123(16):2451-9. doi: 10.1182/blood-2013-08-355818. Epub 2014 Mar 7. Blood. 2014. PMID: 24608975 Free PMC article. Review.
References
-
- Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. - PubMed
-
- Barolo, S., and J. W. Posakony. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167-1181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
