Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination
- PMID: 16287898
- PMCID: PMC1283539
- DOI: 10.1093/nar/gki953
Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination
Abstract
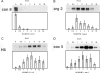
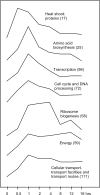
To test the inferences of spotted microarray technology against a biochemically well-studied process, we performed transcriptional profiling of conidial germination in the filamentous fungus, Neurospora crassa. We first constructed a 70 base oligomer microarray that assays 3366 predicted genes. To estimate the relative gene expression levels and changes in gene expression during conidial germination, we analyzed a circuit design of competitive hybridizations throughout a time course using a Bayesian analysis of gene expression level. Remarkable consistency of mRNA profiles with previously published northern data was observed. Genes were hierarchically clustered into groups with respect to their expression profiles over the time course of conidial germination. A functional classification database was employed to characterize the global picture of gene expression. Consensus motif searches identified a putative regulatory component associated with genes involved in ribosomal biogenesis. Our transcriptional profiling data correlate well with biochemical and physiological processes associated with conidial germination and will facilitate functional predictions of novel genes in N.crassa and other filamentous ascomycete species. Furthermore, our dataset on conidial germination allowed comparisons to transcriptional mechanisms associated with germination processes of diverse propagules, such as teliospores of the phytopathogenic fungus Ustilago maydis and spores of the social amoeba Dictyostelium discoideum.
Figures





References
-
- Collins F.S., Lander E.S., Rogers J., Waterston R.H., Conso I.H.G.S. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Davis R.H. NEUROSPORA: Contributions of a Model Organism. NY: Oxford University Press; 2000.
-
- Davis R.H., Perkins D.D. Neurospora: a model of model microbes. Nature Rev. Genet. 2002;3:397–403. - PubMed
-
- Borkovich K.A., Alex L.A., Yarden O., Freitag M., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Molec. Biol. Rev. 2004;68:1–108. - PMC - PubMed
-
- Bistis G.N., Perkins D.D., Read N.D. Cell types of Neurospora crassa. Fungal Genet. Newslett. 2003;50:17–19.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

