Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins
- PMID: 16287971
- PMCID: PMC1297681
- DOI: 10.1073/pnas.0508584102
Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins
Abstract
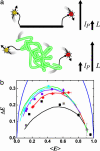
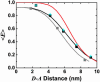
We study protein and nucleic acid structure and dynamics using single-molecule FRET and alternating-laser excitation. Freely diffusing molecules are sorted into subpopulations based on single-molecule signals detected within 100 micros to 1 ms. Distance distributions caused by fluctuations faster than 100 micros are studied within these subpopulations by using time-correlated single-photon counting. Measured distance distributions for dsDNA can be accounted for by considering fluorophore linkers and fluorophore rotational diffusion, except that we find smaller fluctuations for internally labeled dsDNA than DNA with one of the fluorophores positioned at a terminal site. We find that the electrostatic portion of the persistence length of short single-stranded poly(dT) varies approximately as the ionic strength (I) to the -1/2 power (I(-1/2)), and that the average contribution to the contour length per base is 0.40-0.45 nm. We study unfolded chymotrypsin inhibitor 2 (CI2) and unfolded acyl-CoA binding protein (ACBP) even under conditions where folded and unfolded subpopulations coexist (contributions from folded proteins are excluded by using alternating-laser excitation). At lower denaturant concentrations, unfolded CI2 and ACBP are more compact and display larger fluctuations than at higher denaturant concentrations where only unfolded proteins are present. The experimentally measured fluctuations are larger than the fluctuations predicted from a Gaussian chain model or a wormlike chain model. We propose that the larger fluctuations may indicate transient residual structure in the unfolded state.
Figures



References
-
- de Gennes, P. G. (1979) Nature 282, 367–370.
-
- Barrat, J.-L. & Joanny, J.-F. (1996) in Advances in Chemical Physics, eds. Prigogine, I. & Rice, S. A. (Wiley, New York), Vol. 94, pp. 1–65.
-
- Dill, K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10–19. - PubMed
-
- Cannavacciuolo, L. & Pedersen, J. (2002) J. Chem. Phys. 117, 8973–8982.

