MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function
- PMID: 16287976
- PMCID: PMC1287990
- DOI: 10.1073/pnas.0506482102
MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function
Abstract
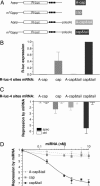
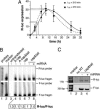
MicroRNAs (miRNAs) repress translation of target mRNAs by interaction with partially mismatched sequences in their 3' UTR. The mechanism by which they act on translation has remained largely obscure. We examined the translation of mRNAs containing four partially mismatched miRNA-binding sites in the 3' UTR in HeLa cells cotransfected with a cognate miRNA. The mRNAs were prepared by in vitro transcription and were engineered to employ different modes of translation initiation. We find that the 5' cap structure and the 3' poly(A) tail are each necessary but not sufficient for full miRNA-mediated repression of mRNA translation. Replacing the cap structure with an internal ribosome entry site from either the cricket paralysis virus or the encephalomyocarditis virus impairs miRNA-mediated repression. Collectively, these results demonstrate that miRNAs interfere with the initiation step of translation and implicate the cap-binding protein eukaryotic initiation factor 4E as a molecular target.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

