The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny
- PMID: 16287978
- PMCID: PMC1283531
- DOI: 10.1073/pnas.0505503102
The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny
Abstract
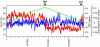
Botulinum neurotoxins (BoNTXs) produced by Clostridium botulinum are among the most poisonous substances known. Of the seven types of BoNTXs, genes for type C1 and D toxins (BoNTX/C1 and D) are carried by bacteriophages. The gene for exoenzyme C3 also resides on these phages. Here, we present the complete genome sequence of c-st, a representative of BoNTX/C1-converting phages. The genome is a linear double-stranded DNA of 185,682 bp with 404-bp terminal direct repeats, the largest known temperate phage genome. We identified 198 potential protein-coding regions, including the genes for production of BoNTX/C1 and exoenzyme C3. Very exceptionally, as a viable bacteriophage, a number of insertion sequences were found on the c-st genome. By analyzing the molecular structure of the c-st genome in lysogens, we also found that it exists as a circular plasmid prophage. These features account for the unstable lysogeny of BoNTX phages, which has historically been called "pseudolysogeny." The PCR scanning analysis of other BoNTX/C1 and D phages based on the c-st sequence further revealed that BoNTX phages comprise a divergent phage family, probably generated by exchanging genomic segments among BoNTX phages and their relatives.
Figures


References
-
- Niemann, H., Blasi, J. & Jahn, R. (1994) Trends Cell Biol. 4, 179-185. - PubMed
-
- Schiavo, G., Matteoli, M. & Montecucco, C. (2000) Physiol. Rev. 80, 717-766. - PubMed
-
- Oguma, K., Inoue, K., Fujinaga, Y., Yokota, K., Watanabe, T., Ohyama, T., Takeshi, K. & Inoue, K. (1999) J. Toxicol. 18, 17-34.
-
- Fujinaga, Y., Inoue, K., Nomura, T., Sasaki, J., Marvaud, J. C., Popoff, M. R., Kozaki, S. & Oguma, K. (2000) FEBS Lett. 467, 179-183. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

