The p53 tumor suppressor network is a key responder to microenvironmental components of chronic inflammatory stress
- PMID: 16288013
- PMCID: PMC1421332
- DOI: 10.1158/0008-5472.CAN-05-1714
The p53 tumor suppressor network is a key responder to microenvironmental components of chronic inflammatory stress
Abstract
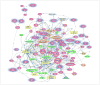
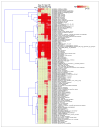
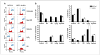
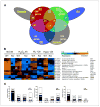
Activation of the p53 network plays a central role in the inflammatory stress response associated with ulcerative colitis and may modulate cancer risk in patients afflicted with this chronic disease. Here, we describe the gene expression profiles associated with four microenvironmental components of the inflammatory response (NO*, H2O2, DNA replication arrest, and hypoxia) that result in p53 stabilization and activation. Isogenic HCT116 and HCT116 TP53-/- colon cancer cells were exposed to the NO* donor Sper/NO, H2O2, hypoxia, or hydroxyurea, and their mRNA was analyzed using oligonucleotide microarrays. Overall, 1,396 genes changed in a p53-dependent manner (P < 0.001), with the majority representing a "unique" profile for each condition. Only 14 genes were common to all four conditions. Included were eight known p53 target genes. Hierarchical sample clustering distinguished early (1 and 4 hours) from late responses (8, 12, and 24 hours), and each treatment was differentiated from the others. Overall, NO* and hypoxia stimulated similar transcriptional responses. Gene ontology analysis revealed cell cycle as a key feature of stress responses and confirmed the similarity between NO* and hypoxia. Cell cycle profiles analyzed by flow cytometry showed that NO* and hypoxia induced quiescent S-phase and G2-M arrest. Using a novel bioinformatic algorithm, we identified several putative p53-responsive elements among the genes induced in a p53-dependent manner, including four [KIAA0247, FLJ12484, p53CSV (HSPC132), and CNK (PLK3)] common to all exposures. In summary, the inflammatory stress response is a complex, integrated biological network in which p53 is a key molecular node regulating gene expression.
Figures


References
-
- Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177–81. - PubMed
-
- Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–85. - PubMed
-
- Li CQ, Robles AI, Hanigan CL, et al. Apoptotic signaling pathways induced by nitric oxide in human lymphoblastoid cells expressing wild-type or mutant p53. Cancer Res. 2004;64:3022–9. - PubMed
-
- Hussain SP, Amstad P, Raja K, et al. Increased p53 mutation load in noncancerous colon tissue fulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Res. 2000;60:3333–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

