HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis
- PMID: 16291646
- PMCID: PMC1283965
- DOI: 10.1101/gad.1338505
HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis
Abstract
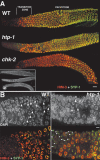
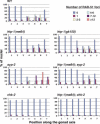
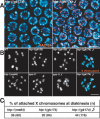
Synaptonemal complex (SC) assembly must occur between correctly paired homologous chromosomes to promote formation of chiasmata. Here, we identify the Caenorhabditis elegans HORMA-domain protein HTP-1 as a key player in coordinating establishment of homolog pairing and synapsis in C. elegans and provide evidence that checkpoint-like mechanisms couple these early meiotic prophase events. htp-1 mutants are defective in the establishment of pairing, but in contrast with the pairing-defective chk-2 mutant, SC assembly is not inhibited and generalized nonhomologous synapsis occurs. Extensive nonhomologous synapsis in htp-1; chk-2 double mutants indicates that HTP-1 is required for the inhibition of SC assembly observed in chk-2 gonads. htp-1 mutants show a decreased abundance of nuclei exhibiting a polarized organization that normally accompanies establishment of pairing; analysis of htp-1; syp-2 double mutants suggests that HTP-1 is needed to prevent premature exit from this polarized nuclear organization and that this exit stops homology search. Further, based on experiments monitoring the formation of recombination intermediates and crossover products, we suggest that htp-1 mutants are defective in preventing the use of sister chromatids as recombination partners. We propose a model in which HTP-1 functions to establish or maintain multiple constraints that operate to ensure coordination of events leading to chiasma formation.
Figures






References
-
- Alpi A., Pasierbek, P., Gartner, A., and Loidl, J. 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. - PubMed
-
- Aravind L. and Koonin, E.V. 1998. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23: 284–286. - PubMed
-
- Armstrong S.J., Caryl, A.P., Jones, G.H., and Franklin, F.C. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
