Increased excision of the Salmonella prophage ST64B caused by a deficiency in Dam methylase
- PMID: 16291663
- PMCID: PMC1291290
- DOI: 10.1128/JB.187.23.7901-7911.2005
Increased excision of the Salmonella prophage ST64B caused by a deficiency in Dam methylase
Abstract
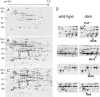
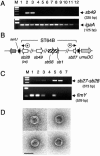
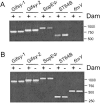
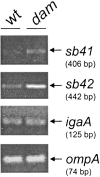
Salmonella enterica mutants defective in Dam methylase are strongly attenuated in virulence and release a large amount of proteins to the extracellular medium. The extent to which these two phenotypes are linked is unknown. Using a proteomic approach, we identified Sb6, Sb13, and Sb36 as proteins present in larger amounts in culture supernatants of an S. enterica serovar Typhimurium dam mutant than in those of the wild-type strain. These three proteins are encoded in the Salmonella prophage ST64B. Higher amounts of ST64B phage DNA and tailless viral capsids were also detected in supernatant extracts of the dam mutant, suggesting that Dam methylation negatively regulates the excision of ST64B. Reverse transcription-PCR analysis revealed that the expression of two ST64B genes encoding a putative antirepressor and a phage replication protein increases in the dam mutant. The SOS response also augments the excision of ST64B. Infection assays performed with phage-cured strains demonstrated that ST64B does not carry genes required for virulence in the mouse model. Evidence was also obtained discarding a relationship between the high excision of ST64B and the envelope instability or virulence attenuation phenotype. Taken together, these data indicate that ST64B excises at a high rate in dam mutants due to the loss of repression exerted by Dam on phage genes and induction of the SOS response characteristic of these mutants. The exacerbated excision of ST64B does not however contribute to the incapacity of dam mutants to cause disease.
Figures

References
-
- Arnold, H. P., U. Ziese, and W. Zillig. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409-416. - PubMed
-
- Baranyi, U., R. Klein, W. Lubitz, D. H. Kruger, and A. Witte. 2000. The archaeal halophilic virus-encoded Dam-like methyltransferase M. phiCh1-I methylates adenine residues and complements dam mutants in the low salt environment of Escherichia coli. Mol. Microbiol. 35:1168-1179. - PubMed
-
- Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

