Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis
- PMID: 16291680
- PMCID: PMC1291260
- DOI: 10.1128/JB.187.23.8063-8080.2005
Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis
Abstract
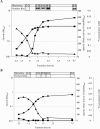
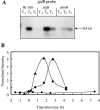
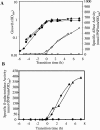
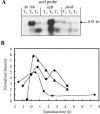
Bacillus subtilis responds to phosphate starvation stress by inducing the PhoP and SigB regulons. While the PhoP regulon provides a specific response to phosphate starvation stress, maximizing the acquisition of phosphate (P(i)) from the environment and reducing the cellular requirement for this essential nutrient, the SigB regulon provides nonspecific resistance to stress by protecting essential cellular components, such as DNA and membranes. We have characterized the phosphate starvation stress response of B. subtilis at a genome-wide level using DNA macroarrays. A combination of outlier and cluster analyses identified putative new members of the PhoP regulon, namely, yfkN (2',3' cyclic nucleotide 2'-phosphodiesterase), yurI (RNase), yjdB (unknown), and vpr (extracellular serine protease). YurI is thought to be responsible for the nonspecific degradation of RNA, while the activity of YfkN on various nucleotide phosphates suggests that it could act on substrates liberated by YurI, which produces 3' or 5' phosphoribonucleotides. The putative new PhoP regulon members are either known or predicted to be secreted and are likely to be important for the recovery of inorganic phosphate from a variety of organic sources of phosphate in the environment.
Figures





References
-
- Akbar, S., S. Y. Lee, S. A. Boylan, and C. W. Price. 1999. Two genes from Bacillus subtilis under the sole control of the general stress transcription factor σB. Microbiology 145:1069-1078. - PubMed
-
- Allenby, N. E., N. O'Connor, Z. Pragai, N. M. Carter, M. Miethke, S. Engelmann, M. Hecker, A. Wipat, A. C. Ward, and C. R. Harwood. 2004. Post-transcriptional regulation of the Bacillus subtilis pst operon encoding a phosphate-specific ABC transporter. Microbiology 150:2619-2628. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

