Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK
- PMID: 16291860
- PMCID: PMC1356581
- DOI: 10.1091/mbc.e05-08-0821
Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK
Abstract
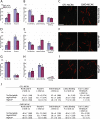
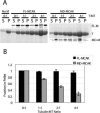
MCAK belongs to the Kinesin-13 family, whose members depolymerize microtubules rather than translocate along them. We defined the minimal functional unit of MCAK as the catalytic domain plus the class specific neck (MD-MCAK), which is consistent with previous reports. We used steady-state ATPase kinetics, microtubule depolymerization assays, and microtubule.MCAK cosedimentation assays to compare the activity of full-length MCAK, which is a dimer, with MD-MCAK, which is a monomer. Full-length MCAK exhibits higher ATPase activity, more efficient microtubule end binding, and reduced affinity for the tubulin heterodimer. Our studies suggest that MCAK dimerization is important for its catalytic cycle by promoting MCAK binding to microtubule ends, enhancing the ability of MCAK to recycle for multiple rounds of microtubule depolymerization, and preventing MCAK from being sequestered by tubulin heterodimers.
Figures

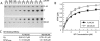



References
-
- Berliner, E., Young, E. C., Anderson, K., Mahtani, H. K., and Gelles, J. (1995). Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature 373, 718-721. - PubMed
-
- Bringmann, H., Skiniotis, G., Spilker, A., Kandels-Lewis, S., Vernos, I., and Surrey, T. (2004). A kinesin-like motor inhibits microtubule dynamic instability. Science 303, 1519-1522. - PubMed
-
- Cassimeris, L., and Spittle, C. (2001). Regulation of microtubule-associated proteins. Int. Rev. Cytol. 210, 163-226. - PubMed
-
- Crevel, I. M., Lockhart, A., and Cross, R. A. (1996). Weak and strong states of kinesin and ncd. J. Mol. Biol. 257, 66-76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

