Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae
- PMID: 16292314
- PMCID: PMC1388271
- DOI: 10.1038/nature04148
Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae
Abstract
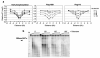
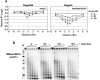
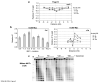
The repair of DNA double-strand breaks (DSBs) is crucial for maintaining genome stability. Eukaryotic cells repair DSBs by both non-homologous end joining and homologous recombination. How chromatin structure is altered in response to DSBs and how such alterations influence DSB repair processes are important issues. In vertebrates, phosphorylation of the histone variant H2A.X occurs rapidly after DSB formation, spreads over megabase chromatin domains, and is required for stable accumulation of repair proteins at damage foci. In Saccharomyces cerevisiae, phosphorylation of the two principal H2A species is also signalled by DSB formation, which spreads approximately 40 kb in either direction from the DSB. Here we show that near a DSB phosphorylation of H2A is followed by loss of histones H2B and H3 and increased sensitivity of chromatin to digestion by micrococcal nuclease; however, phosphorylation of H2A and nucleosome loss occur independently. The DNA damage sensor MRX is required for histone loss, which also depends on INO80, a nucleosome remodelling complex. The repair protein Rad51 (ref. 6) shows delayed recruitment to DSBs in the absence of histone loss, suggesting that MRX-dependent nucleosome remodelling regulates the accessibility of factors directly involved in DNA repair by homologous recombination. Thus, MRX may regulate two pathways of chromatin changes: nucleosome displacement for efficient recruitment of homologous recombination proteins; and phosphorylation of H2A, which modulates checkpoint responses to DNA damage.
Figures

References
-
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. - PubMed
-
- Arkady C, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 2003;5:675–679. - PubMed
-
- Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. - PubMed
-
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

