Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes
- PMID: 16292355
- PMCID: PMC1283159
- DOI: 10.1371/journal.pgen.0010060
Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes
Abstract
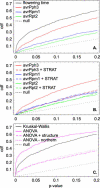
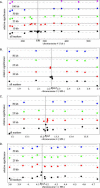
There is currently tremendous interest in the possibility of using genome-wide association mapping to identify genes responsible for natural variation, particularly for human disease susceptibility. The model plant Arabidopsis thaliana is in many ways an ideal candidate for such studies, because it is a highly selfing hermaphrodite. As a result, the species largely exists as a collection of naturally occurring inbred lines, or accessions, which can be genotyped once and phenotyped repeatedly. Furthermore, linkage disequilibrium in such a species will be much more extensive than in a comparable outcrossing species. We tested the feasibility of genome-wide association mapping in A. thaliana by searching for associations with flowering time and pathogen resistance in a sample of 95 accessions for which genome-wide polymorphism data were available. In spite of an extremely high rate of false positives due to population structure, we were able to identify known major genes for all phenotypes tested, thus demonstrating the potential of genome-wide association mapping in A. thaliana and other species with similar patterns of variation. The rate of false positives differed strongly between traits, with more clinal traits showing the highest rate. However, the false positive rates were always substantial regardless of the trait, highlighting the necessity of an appropriate genomic control in association studies.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Nordborg M, Tavaré S. Linkage disequilibrium: What history has to tell us. Trends Genet. 2002;18:83–90. - PubMed
-
- Weiss KM, Terwilliger JD. How many diseases does it take to map a gene with SNPs? Nature Genet. 2000;26:151–157. - PubMed
-
- Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nature Rev Genet. 2004;5:89–100. - PubMed
-
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. - PubMed
-
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, et al. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

