Gene silencing of HIV chemokine receptors using ribozymes and single-stranded antisense RNA
- PMID: 16293105
- PMCID: PMC1408682
- DOI: 10.1042/BJ20051268
Gene silencing of HIV chemokine receptors using ribozymes and single-stranded antisense RNA
Abstract
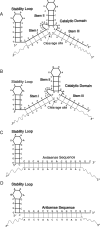
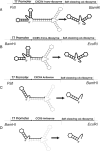
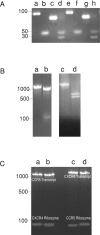
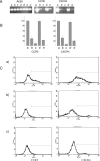
The chemokine receptors CXCR4 and CCR5 are required for HIV-1 to enter cells, and the progression of HIV-1 infection to AIDS involves a switch in the co-receptor usage of the virus from CCR5 to CXCR4. These receptors therefore make attractive candidates for therapeutic intervention, and we have investigated the silencing of their genes by using ribozymes and single-stranded antisense RNAs. In the present study, we demonstrate using ribozymes that a depletion of CXCR4 and CCR5 mRNAs can be achieved simultaneously in human PBMCs (peripheral blood mononuclear cells), cells commonly used by the virus for infection and replication. Ribozyme activity leads to an inhibition of the cell-surface expression of both CCR5 and CXCR4, resulting in a significant inhibition of HIV-1 replication when PBMCs are challenged with the virus. In addition, we show that small single-stranded antisense RNAs can also be used to silence CCR5 and CXCR4 genes when delivered to PBMCs. This silencing is caused by selective degradation of receptor mRNAs.
Figures
Similar articles
-
Characterization of anti-CCR5 ribozyme-transduced CD34+ hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo.Mol Ther. 2000 Mar;1(3):244-54. doi: 10.1006/mthe.2000.0038. Mol Ther. 2000. PMID: 10933940
-
Functional correlation of P-glycoprotein expression and genotype with expression of the human immunodeficiency virus type 1 coreceptor CXCR4.J Virol. 2004 Nov;78(21):12022-9. doi: 10.1128/JVI.78.21.12022-12029.2004. J Virol. 2004. PMID: 15479841 Free PMC article.
-
Inhibition of HIV-1 fusion with small interfering RNAs targeting the chemokine coreceptor CXCR4.Gene Ther. 2004 Dec;11(23):1703-12. doi: 10.1038/sj.gt.3302339. Gene Ther. 2004. PMID: 15306840
-
The application of ribozymes to HIV infection.Curr Opin Mol Ther. 1999 Jun;1(3):316-22. Curr Opin Mol Ther. 1999. PMID: 11713796 Review.
-
CCR5 antagonists: a new tool in fighting HIV.J HIV Ther. 2005 Dec;10(4):68-71. J HIV Ther. 2005. PMID: 16519245 Review.
Cited by
-
Engineering T Cells to Functionally Cure HIV-1 Infection.Mol Ther. 2015 Jul;23(7):1149-1159. doi: 10.1038/mt.2015.70. Epub 2015 Apr 21. Mol Ther. 2015. PMID: 25896251 Free PMC article. Review.
-
Stem cell-based anti-HIV gene therapy.Virology. 2011 Mar 15;411(2):260-72. doi: 10.1016/j.virol.2010.12.039. Epub 2011 Jan 17. Virology. 2011. PMID: 21247612 Free PMC article. Review.
References
-
- Zaitseva M., Peden K., Golding H. HIV coreceptors: role of structure, posttranslational modifications, and internalization in viral-cell fusion and as targets for entry inhibitors. Biochim. Biophys. Acta. 2003;1614:51–61. - PubMed
-
- Bagnarelli P., Fiorelli L., Vecchi M., Monachetti A., Menzo S., Clementi M. Analysis of the functional relationship between V3 loop and gp120 context with regard to human immunodeficiency virus coreceptor usage using naturally selected sequences and different viral backbones. Virology. 2003;307:328–340. - PubMed
-
- Chen B., Vogan E. M., Gong H., Skehel J. J., Wiley D. C., Harrison S. C. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature (London) 2005;433:834–841. - PubMed
-
- Meister S., Otto C., Papkalla A., Krumbiegel M., Pohlmann S., Kirchhoff F. Basic amino acid residues in the V3 loop of simian immunodeficiency virus envelope alter viral coreceptor tropism and infectivity but do not allow efficient utilization of CXCR4 as entry cofactor. Virology. 2001;284:287–296. - PubMed
-
- Samson M., Libert F., Doranz B. J., Rucker J., Liesnard C., Farber C. M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C., et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature (London) 1996;382:722–725. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

