Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway
- PMID: 16293107
- PMCID: PMC1316260
- DOI: 10.1042/BJ20050908
Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway
Abstract
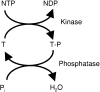
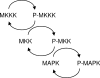
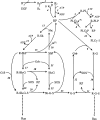
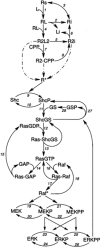
The MAPK (mitogen-activated protein kinase) pathway is one of the most important and intensively studied signalling pathways. It is at the heart of a molecular-signalling network that governs the growth, proliferation, differentiation and survival of many, if not all, cell types. It is de-regulated in various diseases, ranging from cancer to immunological, inflammatory and degenerative syndromes, and thus represents an important drug target. Over recent years, the computational or mathematical modelling of biological systems has become increasingly valuable, and there is now a wide variety of mathematical models of the MAPK pathway which have led to some novel insights and predictions as to how this system functions. In the present review we give an overview of the processes involved in modelling a biological system using the popular approach of ordinary differential equations. Focusing on the MAPK pathway, we introduce the features and functions of the pathway itself before comparing the available models and describing what new biological insights they have led to.
Figures



References
-
- Cobb M. H. MAP kinase pathways. Prog. Biophys. Mol. Biol. 1999;71:479–500. - PubMed
-
- Widmann C., Gibson S., Jarpe M. B., Johnson G. L. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999;79:143–180. - PubMed
-
- Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature (London) 2001;410:37–40. - PubMed
-
- Farooq A., Zhou M. M. Structure and regulation of MAPK phosphatases. Cell. Signalling. 2004;16:769–779. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

