Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes
- PMID: 16298297
- PMCID: PMC1994809
- DOI: 10.1016/j.chembiol.2005.08.011
Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes
Abstract
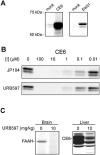
Fatty acid amide hydrolase (FAAH) regulates a large class of signaling lipids, including the endocannabinoid anandamide. Carbamate inhibitors of FAAH display analgesic and anxiolytic properties in rodents. However, the mechanism by which carbamates inhibit FAAH remains obscure. Here, we provide biochemical evidence that carbamates covalently modify the active site of FAAH by adopting an orientation opposite of that originally predicted from modeling. Based on these results, a series of carbamates was designed that display enhanced potency. One agent was converted into a "click chemistry" probe to comprehensively evaluate the proteome reactivity of FAAH-directed carbamates in vivo. These inhibitors were selective for FAAH in the nervous system, but they reacted with several enzymes in peripheral tissues. The experimental strategy described herein can be used to create in vivo probes for any enzyme susceptible to covalent inhibition.
Figures


Comment in
-
Design of on-target FAAH inhibitors.Chem Biol. 2005 Nov;12(11):1157-8. doi: 10.1016/j.chembiol.2005.11.001. Chem Biol. 2005. PMID: 16298293
References
-
- Bos CL, Richel DJ, Ritsema T, Peppelenbosch MP, Versteeg HH. Prostanoids and prostanoid receptors in signal transduction. Int. J. Biochem. Cell Biol. 2004;36:1187–1205. - PubMed
-
- Fukushima N, Ishii I, Contos JJ, Weiner JA, Chun J. Lysophospholipid receptors. Annu. Rev. Pharmacol. Toxicol. 2001;41:507–534. - PubMed
-
- Maccarrone M, Finazzi-Agro A. Endocannabinoids and their actions. Vitam. Horm. 2002;65:225–255. - PubMed
-
- Fowler CJ, Holt S, Nilsson O, Jonsson KO, Tiger G, Jacobsson SO. The endocannabinoid signaling system: pharmacological and therapeutic aspects. Pharmacol. Biochem. Behav. 2005;81:248–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

