Binding-linked protonation of a DNA minor-groove agent
- PMID: 16299076
- PMCID: PMC1367283
- DOI: 10.1529/biophysj.105.071381
Binding-linked protonation of a DNA minor-groove agent
Abstract
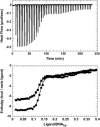
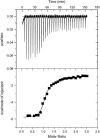
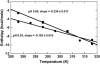
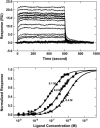
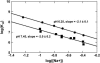
The energetics for binding of a diphenyl diamidine antitrypanosomal agent CGP 40215A to DNA have been studied by spectroscopy, isothermal titration calorimetry, and surface plasmon resonance biosensor methods. Both amidines are positively charged under experimental conditions, but the linking group for the two phenyl amidines has a pK(a) of 6.3 that is susceptible to a protonation process. Spectroscopic studies indicate an increase of 2.7 pK(a) units in the linking group when the compound binds to an A/T minor-groove site. Calorimetric titrations in different buffers and pH conditions support the proton-linkage process and are in a good agreement with spectroscopic titrations. The two methods established a proton-uptake profile as a function of pH. The exothermic enthalpy of complex formation varies with different pH conditions. The observed binding enthalpy increases as a function of temperature indicating a negative heat capacity change that is typical for DNA minor-groove binders. Solvent accessible surface area calculations suggest that surface burial accounts for about one-half of the observed intrinsic negative heat capacity change. Biosensor and calorimetric experiments indicate that the binding affinities vary with pH values and salt concentrations due to protonation and electrostatic interactions. The surface plasmon resonance binding studies indicate that the charge density per phosphate in DNA hairpins is smaller than that in polymers. Energetic contributions from different factors were also estimated for the ligand/DNA complex.
Figures
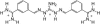


Similar articles
-
Characterization of a novel DNA minor-groove complex.Biophys J. 2004 Feb;86(2):1028-41. doi: 10.1016/s0006-3495(04)74178-8. Biophys J. 2004. PMID: 14747338 Free PMC article.
-
Strong binding in the DNA minor groove by an aromatic diamidine with a shape that does not match the curvature of the groove.J Am Chem Soc. 2002 Nov 20;124(46):13680-1. doi: 10.1021/ja027953c. J Am Chem Soc. 2002. PMID: 12431090
-
Induced fit conformational changes of a "reversed amidine" heterocycle: optimized interactions in a DNA minor groove complex.J Am Chem Soc. 2007 May 2;129(17):5688-98. doi: 10.1021/ja069003n. Epub 2007 Apr 11. J Am Chem Soc. 2007. PMID: 17425312 Free PMC article.
-
Out-of-shape DNA minor groove binders: induced fit interactions of heterocyclic dications with the DNA minor groove.Biochemistry. 2005 Nov 15;44(45):14701-8. doi: 10.1021/bi051791q. Biochemistry. 2005. PMID: 16274217
-
Binding to the DNA minor groove by heterocyclic dications: from AT-specific monomers to GC recognition with dimers.Curr Protoc Nucleic Acid Chem. 2012 Dec;Chapter 8:Unit8.8. doi: 10.1002/0471142700.nc0808s51. Curr Protoc Nucleic Acid Chem. 2012. PMID: 23255206 Free PMC article. Review.
Cited by
-
Targeting RNA by small molecules: comparative structural and thermodynamic aspects of aristololactam-β-D-glucoside and daunomycin binding to tRNA(phe).PLoS One. 2011;6(8):e23186. doi: 10.1371/journal.pone.0023186. Epub 2011 Aug 16. PLoS One. 2011. PMID: 21858023 Free PMC article.
-
Unusually strong binding to the DNA minor groove by a highly twisted benzimidazole diphenylether: induced fit and bound water.Biochemistry. 2007 Jun 12;46(23):6944-56. doi: 10.1021/bi700288g. Epub 2007 May 17. Biochemistry. 2007. PMID: 17506529 Free PMC article.
-
A role for water molecules in DNA-ligand minor groove recognition.Acc Chem Res. 2009 Jan 20;42(1):11-21. doi: 10.1021/ar800016q. Acc Chem Res. 2009. PMID: 18798655 Free PMC article.
-
Thermodynamic profiling of HIV RREIIB RNA-zinc finger interactions.J Mol Biol. 2009 Oct 23;393(2):369-82. doi: 10.1016/j.jmb.2009.07.066. Epub 2009 Jul 30. J Mol Biol. 2009. PMID: 19646998 Free PMC article.
-
Revisiting the association of cationic groove-binding drugs to DNA using a Poisson-Boltzmann approach.Biophys J. 2010 Aug 4;99(3):879-86. doi: 10.1016/j.bpj.2010.04.066. Biophys J. 2010. PMID: 20682266 Free PMC article.
References
-
- Petraccone, L., E. Erra, C. A. Mattia, V. Fedullo, G. Barone, and C. Giancola. 2004. Linkage of proton binding to the thermal dissociation of triple helix complex. Biophys. Chem. 110:73–81. - PubMed
-
- Dullweber, F., M. T. Stubbs, D. Musil, J. Sturzebecher, and G. Klebe. 2001. Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition. J. Mol. Biol. 313:593–614. - PubMed
-
- Li, W., P. Wu, T. Ohmichi, and N. Sugimoto. 2002. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 526:77–81. - PubMed
-
- Jin, E., V. Katritch, W. K. Olson, M. Kharatisvili, R. Abagyan, and D. S. Pilch. 2000. Aminoglycoside binding in the major groove of duplex RNA: the thermodynamic and electrostatic forces that govern recognition. J. Mol. Biol. 298:95–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

