Nucleic acid helix stability: effects of salt concentration, cation valence and size, and chain length
- PMID: 16299077
- PMCID: PMC1367269
- DOI: 10.1529/biophysj.105.070904
Nucleic acid helix stability: effects of salt concentration, cation valence and size, and chain length
Abstract
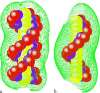
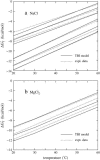
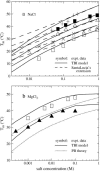
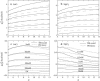
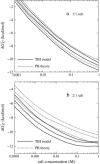
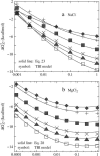
Metal ions play crucial roles in thermal stability and folding kinetics of nucleic acids. For ions (especially multivalent ions) in the close vicinity of nucleic acid surface, interion correlations and ion-binding mode fluctuations may be important. Poisson-Boltzmann theory ignores these effects whereas the recently developed tightly bound ion (TBI) theory explicitly accounts for these effects. Extensive experimental data demonstrate that the TBI theory gives improved predictions for multivalent ions (e.g., Mg2+) than the Poisson-Boltzmann theory. In this study, we use the TBI theory to investigate how the metal ions affect the folding stability of B-DNA helices. We quantitatively evaluate the effects of ion concentration, ion size and valence, and helix length on the helix stability. Moreover, we derive practically useful analytical formulas for the thermodynamic parameters as functions of finite helix length, ion type, and ion concentration. We find that the helix stability is additive for high ion concentration and long helix and nonadditive for low ion concentration and short helix. All these results are tested against and supported by extensive experimental data.
Figures



References
-
- Tinoco, I., and C. Bustamante. 1999. How RNA folds. J. Mol. Biol. 293:271–281. - PubMed
-
- Woodson, S. A. 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 9:104–109. - PubMed
-
- Draper, D. E., D. Grilley, and A. M. Soto. 2005. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34:221–243. - PubMed
-
- Sosnick, T. R., and T. Pan. 2003. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 13:309–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

