Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis
- PMID: 16299223
- PMCID: PMC1315368
- DOI: 10.1105/tpc.105.035261
Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis
Abstract
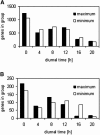
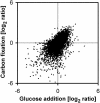
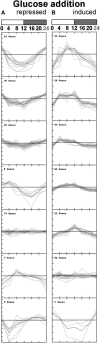
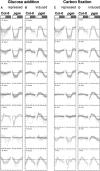
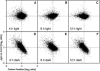
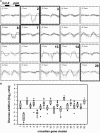
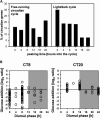
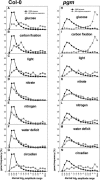
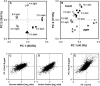
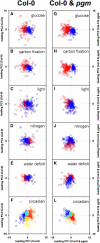
The diurnal cycle strongly influences many plant metabolic and physiological processes. Arabidopsis thaliana rosettes were harvested six times during 12-h-light/12-h-dark treatments to investigate changes in gene expression using ATH1 arrays. Diagnostic gene sets were identified from published or in-house expression profiles of the response to light, sugar, nitrogen, and water deficit in seedlings and 4 h of darkness or illumination at ambient or compensation point [CO(2)]. Many sugar-responsive genes showed large diurnal expression changes, whose timing matched that of the diurnal changes of sugars. A set of circadian-regulated genes also showed large diurnal changes in expression. Comparison of published results from a free-running cycle with the diurnal changes in Columbia-0 (Col-0) and the starchless phosphoglucomutase (pgm) mutant indicated that sugars modify the expression of up to half of the clock-regulated genes. Principle component analysis identified genes that make large contributions to diurnal changes and confirmed that sugar and circadian regulation are the major inputs in Col-0 but that sugars dominate the response in pgm. Most of the changes in pgm are triggered by low sugar levels during the night rather than high levels in the light, highlighting the importance of responses to low sugar in diurnal gene regulation. We identified a set of candidate regulatory genes that show robust responses to alterations in sugar levels and change markedly during the diurnal cycle.
Figures



References
-
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300.
-
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19 185–193. - PubMed
-
- Calikowski, T.T., Meulia, T., and Meier, I. (2003). A proteomic study of the Arabidopsis nuclear matrix. J. Cell. Biochem. 90 361–378. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

