All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote
- PMID: 16299588
- PMCID: PMC1285065
- DOI: 10.1371/journal.pgen.0010067
All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote
Abstract
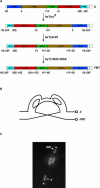
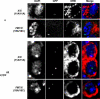
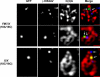
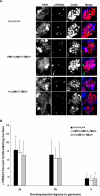
The multiply inverted X chromosome balancer FM7 strongly suppresses, or eliminates, the occurrence of crossing over when heterozygous with a normal sequence homolog. We have utilized the LacI-GFP: lacO system to visualize the effects of FM7 on meiotic pairing, synapsis, and double-strand break formation in Drosophila oocytes. Surprisingly, the analysis of meiotic pairing and synapsis for three lacO reporter couplets in FM7/X heterozygotes revealed they are paired and synapsed during zygotene/pachytene in 70%-80% of oocytes. Moreover, the regions defined by these lacO couplets undergo double-strand break formation at normal frequency. Thus, even complex aberration heterozygotes usually allow high frequencies of meiotic pairing, synapsis, and double-strand break formation in Drosophila oocytes. However, the frequencies of failed pairing and synapsis were still 1.5- to 2-fold higher than were observed for corresponding regions in oocytes with two normal sequence X chromosomes, and this effect was greatest near a breakpoint. We propose that heterozygosity for breakpoints creates a local alteration in synaptonemal complex structure that is propagated across long regions of the bivalent in a fashion analogous to chiasma interference, which also acts to suppress crossing over.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



References
-
- Vazquez J, Belmont AS, Sedat JW. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol. 2002;12:1473–1483. - PubMed
-
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes Dev. 1997;11:2600–2621. - PubMed
-
- Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1997;77:977–991. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

