Translational control by viral proteinases
- PMID: 16303201
- PMCID: PMC7173276
- DOI: 10.1016/j.virusres.2005.10.016
Translational control by viral proteinases
Abstract
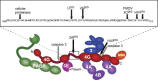
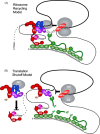
Most RNA viruses have evolved strategies to regulate cellular translation in order to promote preferential expression of the viral genome. Positive strand RNA viruses express large portions, or all of their proteome via translation of large polyproteins that are processed by embedded viral proteinases or host proteinases. Several of these viral proteinases are known to interact with host proteins, particularly with the host translation machinery, and thus, encompass the dual functions of processing of viral polyproteins and exerting translation control. Picornaviruses are perhaps the best characterized in regards to interaction of their proteinases with the host translation machinery and will be emphasized here. However, new findings have shown that similar paradigms exist in other viral systems which will be discussed.
Figures


References
-
- Aldabe R., Feduchi E., Novoa I., Carrasco L. Expression of poliovirus 2Apro in mammalian cells: effects on translation. FEBS Lett. 1995;377(1):1–5. - PubMed
-
- Aminev A.G., Amineva S.P., Palmenberg A.C. Encephalomyocarditis viral protein 2A localizes to nucleoli and inhibits cap-dependent mRNA translation. Virus Res. 2003;95(1–2):45–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

