alphaIIbbeta3 biogenesis is controlled by engagement of alphaIIb in the calnexin cycle via the N15-linked glycan
- PMID: 16304048
- PMCID: PMC1895385
- DOI: 10.1182/blood-2005-07-2990
alphaIIbbeta3 biogenesis is controlled by engagement of alphaIIb in the calnexin cycle via the N15-linked glycan
Abstract
Although much is known about alphaIIbbeta3 structure and function, relatively little is understood about its biogenesis. Thus, we studied the kinetics of pro-alphaIIb production and degradation, focusing on whether proteasomal degradation or the calnexin cycle participates in these processes. In pulse-chase analyses, the time to half-disappearance of pro-alphaIIb (t1/2) was the same in (1) HEK293 cells transfected with (a) alphaIIb plus beta3, (b) alphaIIb alone, (c) mutant V298FalphaIIb plus beta3, or (d) I374TalphaIIb plus beta3; and (2) murine wild-type and beta3-null megakaryocytes. Inhibition of the proteasome prolonged the t1/2 values in both HEK293 cells and murine megakaryocytes. Calnexin coprecipitated with alphaIIb from HEK293 cells transfected with alphaIIb alone, alphaIIb plus beta3, and V298FalphaIIb plus beta3. For proteins in the calnexin cycle, removal of the terminal mannose residue of the middle branch of the core N-linked glycan results in degradation. Inhibition of the enzyme that removes this mannose residue prevented pro-alphaIIb degradation in beta3-null murine megakaryocytes. alphaIIb contains a conserved glycosylation consensus sequence at N15, and an N15Q mutation prevented pro-alphaIIb maturation, complex formation, and degradation. Our findings suggest that pro-alphaIIb engages the calnexin cycle via the N15 glycan and that failure of pro-alphaIIb to complex normally with beta3 results in proteasomal degradation.
Figures

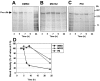



 represents the mannose that is removed by ER mannosidase I. The checkered diamond represents the single glucose in the oligosaccharide structure that is recognized by calnexin. (B) Pulse-chase analyses were performed as described in Figure 1 on bone marrow megakaryocytes from β3-null mice in the presence or absence of the inhibitors 1, deoxy-mannojirimycin (DMJ), deoxynojirimycin (DNJ), or kifunensine (KIF). Whole cell lysates were immunoprecipitated with a combination of mAbs to αIIb (B1B5 and M-148). ER Man I indicates ER mannosidase I; ER Man II, ER mannosidase II; UGGT, UDP-glucose:glycoprotein glucosyltransferase.
represents the mannose that is removed by ER mannosidase I. The checkered diamond represents the single glucose in the oligosaccharide structure that is recognized by calnexin. (B) Pulse-chase analyses were performed as described in Figure 1 on bone marrow megakaryocytes from β3-null mice in the presence or absence of the inhibitors 1, deoxy-mannojirimycin (DMJ), deoxynojirimycin (DNJ), or kifunensine (KIF). Whole cell lysates were immunoprecipitated with a combination of mAbs to αIIb (B1B5 and M-148). ER Man I indicates ER mannosidase I; ER Man II, ER mannosidase II; UGGT, UDP-glucose:glycoprotein glucosyltransferase.
Similar articles
-
AlphaIIbG236E causes Glanzmann thrombasthenia by impairing association with beta3.Platelets. 2008 Aug;19(5):322-7. doi: 10.1080/09537100802123365. Platelets. 2008. PMID: 18791937
-
Integrin alphaIIb promoter-targeted expression of gene products in megakaryocytes derived from retrovirus-transduced human hematopoietic cells.Proc Natl Acad Sci U S A. 1999 Aug 17;96(17):9654-9. doi: 10.1073/pnas.96.17.9654. Proc Natl Acad Sci U S A. 1999. PMID: 10449749 Free PMC article.
-
Two novel mutations in the alpha IIb calcium-binding domains identify hydrophobic regions essential for alpha IIbbeta 3 biogenesis.Blood. 2003 Mar 15;101(6):2268-76. doi: 10.1182/blood-2002-07-2266. Epub 2002 Nov 7. Blood. 2003. PMID: 12424194
-
Cytoplasmic binding partners of the platelet integrin alphaIIb beta3.Front Biosci. 2007 Jan 1;12:2038-49. doi: 10.2741/2209. Front Biosci. 2007. PMID: 17127442 Review.
-
Platelet glycoprotein IIb/IIIa antagonists: lessons learned from clinical trials and future directions.Crit Care Med. 2002 May;30(5 Suppl):S332-40. doi: 10.1097/00003246-200205001-00025. Crit Care Med. 2002. PMID: 12004256 Review.
Cited by
-
Beyond COX-1: the effects of aspirin on platelet biology and potential mechanisms of chemoprevention.Cancer Metastasis Rev. 2017 Jun;36(2):289-303. doi: 10.1007/s10555-017-9675-z. Cancer Metastasis Rev. 2017. PMID: 28762014 Free PMC article. Review.
-
Asn54-linked glycan is critical for functional folding of intercellular adhesion molecule-5.Glycoconj J. 2012 Jan;29(1):47-55. doi: 10.1007/s10719-011-9363-0. Epub 2011 Dec 21. Glycoconj J. 2012. PMID: 22187327
-
In silico analysis of structural modifications in and around the integrin αIIb genu caused by ITGA2B variants in human platelets with emphasis on Glanzmann thrombasthenia.Mol Genet Genomic Med. 2018 Mar;6(2):249-260. doi: 10.1002/mgg3.365. Epub 2018 Jan 31. Mol Genet Genomic Med. 2018. PMID: 29385657 Free PMC article.
-
Integrating Phenotypic and Chemoproteomic Approaches to Identify Covalent Targets of Dietary Electrophiles in Platelets.ACS Cent Sci. 2024 Jan 29;10(2):344-357. doi: 10.1021/acscentsci.3c00822. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435523 Free PMC article.
-
Molecular dynamics simulations involving different β-propeller mutations reported in Swiss and French patients correlate with their disease phenotypes.Sci Rep. 2024 Oct 15;14(1):24133. doi: 10.1038/s41598-024-75070-4. Sci Rep. 2024. PMID: 39406775 Free PMC article.
References
-
- Phillips DR, Charo IF, Parise LV, Fitzgerald LA. The platelet membrane glycoprotein IIb-IIIa complex. Blood. 1988;71: 831-843. - PubMed
-
- George JN, Caen JP, Nurden AT. Glanzmann's thrombasthenia: the spectrum of clinical disease. Blood. 1990;75: 1383-1395. - PubMed
-
- Rosa J-P, McEver RP. Processing and assembly of the integrin, glycoprotein IIb-IIIa, in HEL cells. J Biol Chem. 1989;264: 12596-12603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

