AntiJen: a quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data
- PMID: 16305757
- PMCID: PMC1289288
- DOI: 10.1186/1745-7580-1-4
AntiJen: a quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data
Abstract
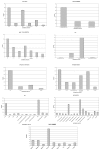
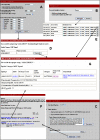
AntiJen is a database system focused on the integration of kinetic, thermodynamic, functional, and cellular data within the context of immunology and vaccinology. Compared to its progenitor JenPep, the interface has been completely rewritten and redesigned and now offers a wider variety of search methods, including a nucleotide and a peptide BLAST search. In terms of data archived, AntiJen has a richer and more complete breadth, depth, and scope, and this has seen the database increase to over 31,000 entries. AntiJen provides the most complete and up-to-date dataset of its kind. While AntiJen v2.0 retains a focus on both T cell and B cell epitopes, its greatest novelty is the archiving of continuous quantitative data on a variety of immunological molecular interactions. This includes thermodynamic and kinetic measures of peptide binding to TAP and the Major Histocompatibility Complex (MHC), peptide-MHC complexes binding to T cell receptors, antibodies binding to protein antigens and general immunological protein-protein interactions. The database also contains quantitative specificity data from position-specific peptide libraries and biophysical data, in the form of diffusion co-efficients and cell surface copy numbers, on MHCs and other immunological molecules. The uses of AntiJen include the design of vaccines and diagnostics, such as tetramers, and other laboratory reagents, as well as helping parameterize the bioinformatic or mathematical in silico modeling of the immune system. The database is accessible from the URL: http://www.jenner.ac.uk/antijen.
Figures

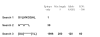
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
