Sequence, distance, and accessibility are determinants of 5'-end-directed cleavages by retroviral RNases H
- PMID: 16306040
- PMCID: PMC1360142
- DOI: 10.1074/jbc.M510504200
Sequence, distance, and accessibility are determinants of 5'-end-directed cleavages by retroviral RNases H
Abstract
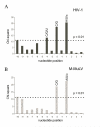
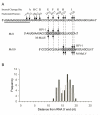
The RNase H activity of reverse transcriptase is essential for retroviral replication. RNA 5'-end-directed cleavages represent a form of RNase H activity that is carried out on RNA/DNA hybrids that contain a recessed RNA 5'-end. Previously, the distance from the RNA 5'-end has been considered the primary determinant for the location of these cleavages. Employing model hybrid substrates and the HIV-1 and Moloney murine leukemia virus reverse transcriptases, we demonstrate that cleavage sites correlate with specific sequences and that the distance from the RNA 5'-end determines the extent of cleavage. An alignment of sequences flanking multiple RNA 5'-end-directed cleavage sites reveals that both enzymes strongly prefer A or U at the +1 position and C or G at the -2 position, and additionally for HIV-1, A is disfavored at the -4 position. For both enzymes, 5'-end-directed cleavages occurred when sites were positioned between the 13th and 20th nucleotides from the RNA 5'-end, a distance termed the cleavage window. In examining the importance of accessibility to the RNA 5'-end, it was found that the extent of 5'-end-directed cleavages observed in substrates containing a free recessed RNA 5'-end was most comparable to substrates with a gap of two or three bases between the upstream and downstream RNAs. Together these finding demonstrate that the selection of 5'-end-directed cleavage sites by retroviral RNases H results from a combination of nucleotide sequence, permissible distance, and accessibility to the RNA 5'-end.
Figures









References
-
- Arts EJ, LeGrice SFJ. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:339–393. - PubMed
-
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, N. Y.: 1997. - PubMed
-
- di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Science. 1986;231:1289–1291. - PubMed
-
- Misra HS, Pandey PK, Pandey VN. J. Biol. Chem. 1998;273:9785–9789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

