Subcellular localization and light-regulated expression of protoporphyrinogen IX oxidase and ferrochelatase in Chlamydomonas reinhardtii
- PMID: 16306143
- PMCID: PMC1310572
- DOI: 10.1104/pp.105.069732
Subcellular localization and light-regulated expression of protoporphyrinogen IX oxidase and ferrochelatase in Chlamydomonas reinhardtii
Abstract
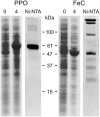
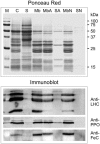
Protoporphyrinogen IX oxidase (PPO) catalyzes the last common step in chlorophyll and heme synthesis, and ferrochelatase (FeC) catalyzes the last step of the heme synthesis pathway. In plants, each of these two enzymes is encoded by two or more genes, and the enzymes have been reported to be located in the chloroplasts or in the mitochondria. We report that in the green alga Chlamydomonas reinhardtii, PPO and FeC are each encoded by a single gene. Phylogenetic analysis indicates that C. reinhardtii PPO and FeC are most closely related to plant counterparts that are located only in chloroplasts. Immunoblotting results suggest that C. reinhardtii PPO and FeC are targeted exclusively to the chloroplast, where they are associated with membranes. These results indicate that cellular needs for heme in this photosynthetic eukaryote can be met by heme that is synthesized in the chloroplast. It is proposed that the multiplicity of genes for PPO and FeC in higher plants could be related to differential expression in differently developing tissues rather than to targeting of different gene products to different organelles. The FeC content is higher in C. reinhardtii cells growing in continuous light than in cells growing in the dark, whereas the content of PPO does not significantly differ in light- and dark-grown cells. In cells synchronized to a light/dark cycle, the level of neither enzyme varied significantly with the phase of the cycle. These results indicate that heme synthesis is not directly regulated by the levels of PPO and FeC in C. reinhardtii.
Figures





References
-
- Armbrust EV, Ferris PJ, Goodenough UW (1993) A mating-type linked gene-cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell 74: 801–811 - PubMed
-
- Beale SI (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60: 43–73
-
- Beale SI (2005) Green genes gleaned. Trends Plant Sci 10: 309–312 - PubMed
-
- Chow KS, Singh DP, Walker AR, Smith AG (1998) Two different genes encode ferrochelatase in Arabidopsis: mapping, expression and subcellular targeting of the precursor proteins. Plant J 15: 531–541 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

