Bacterial DNA segregation by dynamic SopA polymers
- PMID: 16306264
- PMCID: PMC1308903
- DOI: 10.1073/pnas.0507222102
Bacterial DNA segregation by dynamic SopA polymers
Abstract
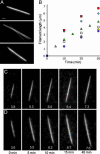
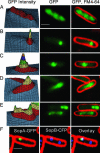
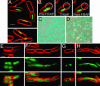
Many bacterial plasmids and chromosomes rely on ParA ATPases for proper positioning within the cell and for efficient segregation to daughter cells. Here we demonstrate that the F-plasmid-partitioning protein SopA polymerizes into filaments in an ATP-dependent manner in vitro, and that the filaments elongate at a rate that is similar to that of plasmid separation in vivo. We show that SopA is a dynamic protein within the cell, undergoing cycles of polymerization and depolymerization, and shuttling back and forth between nucleoprotein complexes that are composed of the SopB protein bound to sopC-containing plasmids (SopB/sopC). The dynamic behavior of SopA is critical for Sop-mediated plasmid DNA segregation; mutations that lock SopA into a static polymer in the cell inhibit plasmid segregation. We show that SopA colocalizes with SopB/sopC in the cell and that SopB/sopC nucleates the assembly of SopA and is required for its dynamic behavior. When SopA is polymerized in vitro in the presence of SopB and sopC-containing DNA, SopA filaments emanate from the plasmid DNA in radial asters. We propose a mechanism in which plasmid separation is driven by the polymerization of SopA, and we speculate that the radial assembly of SopA polymers is responsible for positioning plasmids both before and after segregation.
Figures



References
-
- Gadde, S. & Heald, R. (2004) Curr. Biol. 14, R797–R805. - PubMed
-
- Kline-Smith, S. L. & Walczak, C. E. (2004) Mol. Cell 15, 317–327. - PubMed
-
- Gitai, Z., Thanbichler, M. & Shapiro, L. (2005) Trends Microbiol. 13, 221–228. - PubMed
-
- Gerdes, K., Møller-Jensen, J., Ebersbach, G., Kruse, T. & Nordström, K. (2004) Cell 116, 359–366. - PubMed
-
- Thanbichler, M., Viollier, P. H. & Shapiro, L. (2005) Curr. Opin. Genet. Dev. 15, 153–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

