The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme
- PMID: 16306591
- PMCID: PMC1316033
- DOI: 10.1128/JVI.79.24.15199-15208.2005
The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme
Abstract
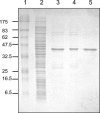
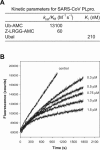
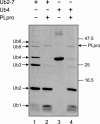
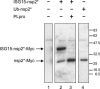
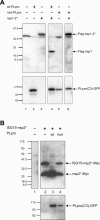
The severe acute respiratory syndrome coronavirus papain-like protease (SARS-CoV PLpro) is involved in the processing of the viral polyprotein and, thereby, contributes to the biogenesis of the virus replication complex. Structural bioinformatics has revealed a relationship for the SARS-CoV PLpro to herpesvirus-associated ubiquitin-specific protease (HAUSP), a ubiquitin-specific protease, indicating potential deubiquitinating activity in addition to its function in polyprotein processing (T. Sulea, H. A. Lindner, E. O. Purisima, and R. Menard, J. Virol. 79:4550-4551, 2005). In order to confirm this prediction, we overexpressed and purified SARS-CoV PLpro (amino acids [aa]1507 to 1858) from Escherichia coli. The purified enzyme hydrolyzed ubiquitin-7-amino-4-methylcoumarin (Ub-AMC), a general deubiquitinating enzyme substrate, with a catalytic efficiency of 13,100 M(-1)s(-1), 220-fold more efficiently than the small synthetic peptide substrate Z-LRGG-AMC, which incorporates the C-terminal four residues of ubiquitin. In addition, SARS-CoV PLpro was inhibited by the specific deubiquitinating enzyme inhibitor ubiquitin aldehyde, with an inhibition constant of 210 nM. The purified SARS-CoV PLpro disassembles branched polyubiquitin chains with lengths of two to seven (Ub2-7) or four (Ub4) units, which involves isopeptide bond cleavage. SARS-CoV PLpro processing activity was also detected against a protein fused to the C terminus of the ubiquitin-like modifier ISG15, both in vitro using the purified enzyme and in HeLa cells by coexpression with SARS-CoV PLpro (aa 1198 to 2009). These results clearly establish that SARS-CoV PLpro is a deubiquitinating enzyme, thereby confirming our earlier prediction. This unexpected activity for a coronavirus papain-like protease suggests a novel viral strategy to modulate the host cell ubiquitination machinery to its advantage.
Figures

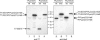
References
-
- Amerik, A. Y., and M. Hochstrasser. 2004. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695:189-207. - PubMed
-
- Andres, G., A. Alejo, C. Simon-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. - PubMed
-
- Baker, R. T. 1996. Protein expression using ubiquitin fusion and cleavage. Curr. Opin. Biotechnol. 7:541-546. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

