Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy
- PMID: 16306598
- PMCID: PMC1316018
- DOI: 10.1128/JVI.79.24.15265-15276.2005
Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy
Abstract
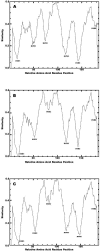
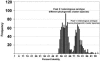
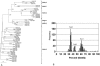
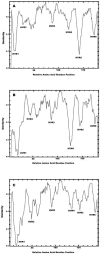
Human adenoviruses (HAdV) are responsible for a wide spectrum of diseases. The neutralization epsilon determinant (loops 1 and 2) and the hemagglutination gamma determinant are relevant for the taxonomy of HAdV. Precise type identification of HAdV prototypes is crucial for detection of infection chains and epidemiology. epsilon and gamma determinant sequences of all 51 HAdV were generated to propose molecular classification criteria. Phylogenetic analysis of epsilon determinant sequences demonstrated sufficient genetic divergence for molecular classification, with the exception of HAdV-15 and HAdV-29, which also cannot be differentiated by classical cross-neutralization. Precise sequence divergence criteria for typing (<2.5% from loop 2 prototype sequence and <2.4% from loop 1 sequence) were deduced from phylogenetic analysis. These criteria may also facilitate identification of new HAdV prototypes. Fiber knob (gamma determinant) phylogeny indicated a two-step model of species evolution and multiple intraspecies recombination events in the origin of HAdV prototypes. HAdV-29 was identified as a recombination variant of HAdV-15 (epsilon determinant) and a speculative, not-yet-isolated HAdV prototype (gamma determinant). Subanalysis of molecular evolution in hypervariable regions 1 to 6 of the epsilon determinant indicated different selective pressures in subclusters of species HAdV-D. Additionally, gamma determinant phylogenetic analysis demonstrated that HAdV-8 did not cluster with -19 and -37 in spite of their having the same tissue tropism. The phylogeny of HAdV-E4 suggested origination by interspecies recombination between HAdV-B (hexon) and HAdV-C (fiber), as in simian adenovirus 25, indicating additional zoonotic transfer. In conclusion, molecular classification by systematic sequence analysis of immunogenic determinants yields new insights into HAdV phylogeny and evolution.
Figures



References
-
- Adrian, T., B. Bastian, W. Benoist, J. C. Hierholzer, and R. Wigand. 1985. Characterization of adenovirus 15/H9 intermediate strains. Intervirology 23:15-22. - PubMed
-
- Adrian, T., G. Wadell, J. C. Hierholzer, and R. Wiegand. 1986. DNA restriction analysis of adenovirus prototype 1 to 41. Arch. Virol. 91:277-290. - PubMed
-
- Akiyama, H., T. Kurosu, C. Sakashita, T. Inoue, S. Mori, K. Ohashi, S. Tanikawa, H. Sakamaki, Y. Onozawa, Q. Chen, H. Zheng, and T. Kitamura. 2001. Adenovirus is a key pathogen in hemorrhagic cystitis associated with bone marrow transplantation. Clin. Infect. Dis. 32:1325-1330. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

