Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein e2 contribute to virus entry
- PMID: 16306604
- PMCID: PMC1316016
- DOI: 10.1128/JVI.79.24.15331-15341.2005
Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein e2 contribute to virus entry
Abstract
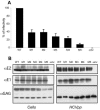
The N terminus of hepatitis C virus (HCV) envelope glycoprotein E2 contains a hypervariable region (HVR1) which has been proposed to play a role in viral entry. Despite strong amino acid variability, HVR1 is globally basic, with basic residues located at specific sequence positions. Here we show by analyzing a large number of HVR1 sequences that the frequency of basic residues at each position is genotype dependent. We also used retroviral pseudotyped particles (HCVpp) harboring genotype 1a envelope glycoproteins to study the role of HVR1 basic residues in entry. Interestingly, HCVpp infectivity globally increased with the number of basic residues in HVR1. However, a shift in position of some charged residues also modulated HCVpp infectivity. In the absence of basic residues, infectivity was reduced to the same level as that of a mutant deleted of HVR1. We also analyzed the effect of these mutations on interactions with some potential HCV receptors. Recognition of CD81 was not affected by changes in the number of charged residues, and we did not find a role for heparan sulfates in HCVpp entry. The involvement of the scavenger receptor class B type I (SR-BI) was indirectly analyzed by measuring the enhancement of infectivity of the mutants in the presence of the natural ligand of SR-BI, high-density lipoproteins (HDL). However, no correlation between the number of basic residues within HVR1 and HDL enhancement effect was observed. Despite the lack of evidence of the involvement of known potential receptors, our results demonstrate that the presence of basic residues in HVR1 facilitates virus entry.
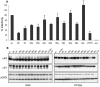
Figures



References
-
- Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. - PubMed
-
- Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between the hyper-variable region 1 of the HCV E2 glycoprotein, the scavenger receptor BI and HDL promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. - PMC - PubMed
-
- Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

