Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7
- PMID: 16306619
- PMCID: PMC1316045
- DOI: 10.1128/JVI.79.24.15477-15493.2005
Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7
Abstract
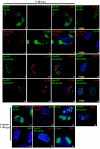
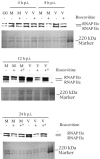
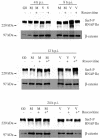
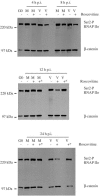
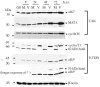
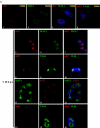
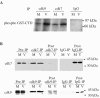
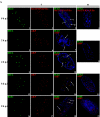
Human cytomegalovirus infection in the presence of the cyclin-dependent kinase (cdk) inhibitor roscovitine leads to changes in differential splicing and the polyadenylation of immediate early IE1/IE2 and UL37 transcripts (V. Sanchez, A. K. McElroy, J. Yen, S. Tamrakar, C. L. Clark, R. A. Schwartz, and D. H. Spector, J. Virol. 78:11219-11232, 2004). To determine if this was associated with specific phosphorylation of the C-terminal domain (CTD) of the RNA polymerase II (RNAP II) large subunit by cdk7/cyclin H and cdk9/cyclin T1, we examined the expression and localization of these kinases and the various phosphorylated forms of RNAP II. Infection resulted in increased RNAP II CTD phosphorylated on serines 2 and 5 and increased levels of activity of cdk7 and cdk9. At early times, cdk9 localizes with input viral DNA, and aggregates of cdk9 and cdk7 and a subset of Ser2-phosphorylated RNAP II colocalize with IE1/IE2 proteins adjacent to promyelocytic leukemia protein oncogenic domains. Later, cdk9 and Ser2-phosphorylated RNAP II form a nuclear punctate pattern; cdk7 resides in replication centers, and Ser5-phosphorylated RNAP II clusters at the peripheries of replication centers. Roscovitine treatment leads to decreased levels of hyperphosphorylated RNAP II (RNAP IIo) in infected cells and of hypophosphorylated RNAP II in mock-infected and infected cells. The RNAP IIo decrease does not occur if roscovitine is added 8 h postinfection, as was previously observed for processing of IE transcripts. These results suggest that accurate IE gene expression requires specific phosphorylation of the RNAP II CTD early in infection.
Figures




Similar articles
-
Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome.J Virol. 2008 Jan;82(1):394-407. doi: 10.1128/JVI.01681-07. Epub 2007 Oct 17. J Virol. 2008. PMID: 17942543 Free PMC article.
-
Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription.Mol Cell Biol. 2000 Jul;20(14):5077-86. doi: 10.1128/MCB.20.14.5077-5086.2000. Mol Cell Biol. 2000. PMID: 10866664 Free PMC article.
-
Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs.BMC Genomics. 2004 Sep 20;5:69. doi: 10.1186/1471-2164-5-69. BMC Genomics. 2004. PMID: 15380029 Free PMC article.
-
CDK9 keeps RNA polymerase II on track.Cell Mol Life Sci. 2021 Jul;78(14):5543-5567. doi: 10.1007/s00018-021-03878-8. Epub 2021 Jun 19. Cell Mol Life Sci. 2021. PMID: 34146121 Free PMC article. Review.
-
The emerging picture of CDK9/P-TEFb: more than 20 years of advances since PITALRE.Mol Biosyst. 2017 Jan 31;13(2):246-276. doi: 10.1039/c6mb00387g. Mol Biosyst. 2017. PMID: 27833949 Review.
Cited by
-
Cyclin-dependent kinase activity controls the onset of the HCMV lytic cycle.PLoS Pathog. 2010 Sep 9;6(9):e1001096. doi: 10.1371/journal.ppat.1001096. PLoS Pathog. 2010. PMID: 20844576 Free PMC article.
-
Viral serine/threonine protein kinases.J Virol. 2011 Feb;85(3):1158-73. doi: 10.1128/JVI.01369-10. Epub 2010 Nov 17. J Virol. 2011. PMID: 21084474 Free PMC article. Review.
-
Combined Treatment with Host-Directed and Anticytomegaloviral Kinase Inhibitors: Mechanisms, Synergisms and Drug Resistance Barriers.Pharmaceutics. 2023 Nov 27;15(12):2680. doi: 10.3390/pharmaceutics15122680. Pharmaceutics. 2023. PMID: 38140021 Free PMC article.
-
Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication.PLoS Pathog. 2007 Jan;3(1):e6. doi: 10.1371/journal.ppat.0030006. PLoS Pathog. 2007. PMID: 17206862 Free PMC article.
-
Human cytomegalovirus and Herpes Simplex type I virus can engage RNA polymerase I for transcription of immediate early genes.Oncotarget. 2017 Oct 29;8(57):96536-96552. doi: 10.18632/oncotarget.22106. eCollection 2017 Nov 14. Oncotarget. 2017. PMID: 29228551 Free PMC article.
References
-
- Adair, R., G. W. Liebisch, and A. M. Colberg-Poley. 2003. Complex alternative processing of human cytomegalovirus UL37 pre-mRNA. J. Gen. Virol. 84:3353-3358. - PubMed
-
- Adair, R., G. W. Liebisch, Y. Su, and A. M. Colberg-Poley. 2004. Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J. Gen. Virol. 85:3542-3553. - PubMed
-
- Ahn, J.-H., W.-J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. - PMC - PubMed
-
- Ahn, J. H., E. R. Brignole, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

