Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses
- PMID: 16306628
- PMCID: PMC1315997
- DOI: 10.1128/JVI.79.24.15573-15577.2005
Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses
Abstract
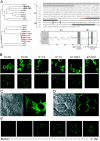
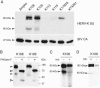
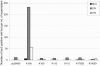
Genome-wide screening of sequence databases for human endogenous retroviruses (HERVs) has led to the identification of 18 coding env genes, among which two-the syncytin genes-encode fusogenic ENV proteins possibly involved in placenta physiology. Here we show that a third ENV, originating from the most "recent" HERV-K(HML2) family, is functional. Immunofluorescence analysis of env-transduced cells demonstrates expression of the protein at the cell surface, and we show that the protein confers infectivity to simian immunodeficiency virus pseudotypes. Western blot analysis of the pseudotyped virions further discloses the expected specific cleavage of the ENV precursor protein. This functional ENV could play a role in the amplification--via infection of the germ line--of the HERV-K genomic copies, all the more as coding HERV-K gag and pol genes can similarly be found in the human genome, which could therefore generate infectious virions of a fully endogenous origin.
Figures
References
-
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

