Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome
- PMID: 16307924
- PMCID: PMC2631382
- DOI: 10.1016/j.molcel.2005.10.007
Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome
Abstract
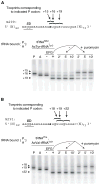
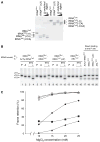
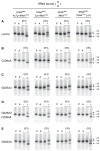
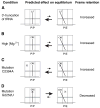
Retention of the reading frame in ribosomal complexes after single-round translocation depends on the acylation state of the tRNA. When tRNA lacking a peptidyl group is translocated to the P site, the mRNA slips to allow re-pairing of the tRNA with a nearby out-of-frame codon. Here, we show that this ribosomal activity results from movement of tRNA into the P/E hybrid state. Slippage of mRNA is suppressed by 3' truncation of the translocated tRNA, increased MgCl2 concentration, and mutation C2394A of the 50S E site, and each of these conditions inhibits P/E-state formation. Mutation G2252U of the 50S P site stimulates mRNA slippage, suggesting that decreased affinity of tRNA for the P/P state also destabilizes mRNA in the complex. The effects of G2252U are suppressed by C2394A, further implicating the P/E state in mRNA destabilization. This work uncovers a functional attribute of the P/E state crucial for understanding translation.
Figures


Similar articles
-
Binding of misacylated tRNAs to the ribosomal A site.RNA. 2005 Nov;11(11):1610-5. doi: 10.1261/rna.2130505. RNA. 2005. PMID: 16244128 Free PMC article.
-
Destabilization of codon-anticodon interaction in the ribosomal exit site.J Mol Biol. 1987 Jul 5;196(1):137-48. doi: 10.1016/0022-2836(87)90516-x. J Mol Biol. 1987. PMID: 2443714
-
Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA.J Mol Biol. 2001 Jun 22;309(5):1029-48. doi: 10.1006/jmbi.2001.4717. J Mol Biol. 2001. PMID: 11399077
-
[tRNA-binding centers of Escherichia coli ribosomes and their structural organization].Mol Biol (Mosk). 1984 Sep-Oct;18(5):1194-207. Mol Biol (Mosk). 1984. PMID: 6209546 Review. Russian.
-
[Mechanism of the stereospecific stabilization of codon-anticodon complexes in ribosomes during translation].Zh Obshch Biol. 1985 Jan-Feb;46(1):63-77. Zh Obshch Biol. 1985. PMID: 3885616 Review. Russian. No abstract available.
Cited by
-
Contribution of an alternative 16S rRNA helix to biogenesis of the 30S subunit of the ribosome.RNA. 2024 Jun 17;30(7):770-778. doi: 10.1261/rna.079960.124. RNA. 2024. PMID: 38570183 Free PMC article.
-
tRNA-mRNA mimicry drives translation initiation from a viral IRES.Nat Struct Mol Biol. 2008 Jan;15(1):57-64. doi: 10.1038/nsmb1351. Epub 2007 Dec 23. Nat Struct Mol Biol. 2008. PMID: 18157151 Free PMC article.
-
Structural basis for mRNA and tRNA positioning on the ribosome.Proc Natl Acad Sci U S A. 2006 Oct 24;103(43):15830-4. doi: 10.1073/pnas.0607541103. Epub 2006 Oct 12. Proc Natl Acad Sci U S A. 2006. PMID: 17038497 Free PMC article.
-
Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs.Mol Cell. 2015 Mar 19;57(6):1059-1073. doi: 10.1016/j.molcel.2015.01.039. Mol Cell. 2015. PMID: 25794616 Free PMC article.
-
Reinitiation and other unconventional posttermination events during eukaryotic translation.Mol Cell. 2013 Jul 25;51(2):249-64. doi: 10.1016/j.molcel.2013.05.026. Epub 2013 Jun 27. Mol Cell. 2013. PMID: 23810859 Free PMC article.
References
-
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. Effect of buffer conditions on the position of tRNA on the 70S ribosome as visualized by cryoelectron microscopy. J Biol Chem. 1999;274:8723–8729. - PubMed
-
- Ehrenberg M, Bilgin N, Kurland CG. Design and use of a fast and accurate in vitro translation system. In: Spedding G, editor. Ribosomes and Protein Synthesis- A Practical Approach. Oxford: IRL Press; 1990. pp. 101–129.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

