A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA
- PMID: 16308566
- PMCID: PMC1356319
- DOI: 10.1038/sj.emboj.7600880
A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA
Abstract
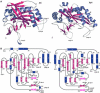
Many reactions in cells proceed via the sequestration of two DNA molecules in a synaptic complex. SfiI is a member of a growing family of restriction enzymes that can bind and cleave two DNA sites simultaneously. We present here the structures of tetrameric SfiI in complex with cognate DNA. The structures reveal two different binding states of SfiI: one with both DNA-binding sites fully occupied and the other with fully and partially occupied sites. These two states provide details on how SfiI recognizes and cleaves its target DNA sites, and gives insight into sequential binding events. The SfiI recognition sequence (GGCCNNNN[downward arrow]NGGCC) is a subset of the recognition sequence of BglI (GCCNNNN[downward arrow]NGGC), and both enzymes cleave their target DNAs to leave 3-base 3' overhangs. We show that even though SfiI is a tetramer and BglI is a dimer, and there is little sequence similarity between the two enzymes, their modes of DNA recognition are unusually similar.
Figures
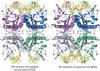
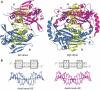


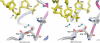
References
-
- Aggarwal AK (1995) Structure and function of restriction endonucleases. Curr Opin Struct Biol 5: 11–19 - PubMed
-
- Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC (1988) Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242: 899–907 - PubMed
-
- Alves J, Ruter T, Geiger R, Fliess A, Maass G, Pingoud A (1989) Changing the hydrogen-bonding potential in the DNA binding site of EcoRI by site-directed mutagenesis drastically reduces the enzymatic activity, not, however, the preference of this restriction endonuclease for cleavage within the site-GAATTC. Biochemistry 28: 2678–2684 - PubMed
-
- Alves J, Vennekohl P (2004) Protein engineering of restriction enzymes. In Restriction Endonucleases, Pingoud A (ed), Vol 14, pp 393–411. Berlin, Heidelberg: Springer-Verlag
-
- Bath AJ, Milsom SE, Gormley NA, Halford SE (2002) Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J Biol Chem 277: 4024–4033 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

