Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability
- PMID: 16314296
- PMCID: PMC1297710
- DOI: 10.1093/nar/gni181
Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability
Abstract
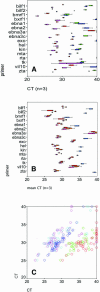
Quantitative real-time PCR has become the method of choice for measuring mRNA transcription. Recently, fast PCR protocols have been developed as a means to increase assay throughput. Yet it is unclear whether more rapid cycling conditions preserve the original assay performance characteristics. We compared 16 primer sets directed against Epstein-Barr virus (EBV) mRNAs using universal and fast PCR cycling conditions. These primers are of clinical relevance, since they can be used to monitor viral oncogene and drug-resistance gene expression in transplant patients and EBV-associated cancers. While none of the primers failed under fast PCR conditions, the fast PCR protocols performed worse than universal cycling conditions. Fast PCR was associated with a loss of sensitivity as well as higher variability, but not with a loss of specificity or with a higher false positive rate.
Figures




References
-
- Dittmer D.P. Transcription profile of Kaposi's sarcoma-associated herpesvirus in primary Kaposi's sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 2003;63:2010–2015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

