Real-time quantification of microRNAs by stem-loop RT-PCR
- PMID: 16314309
- PMCID: PMC1292995
- DOI: 10.1093/nar/gni178
Real-time quantification of microRNAs by stem-loop RT-PCR
Abstract
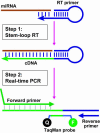
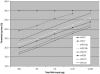
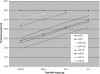
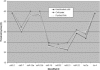
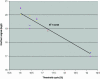
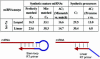
A novel microRNA (miRNA) quantification method has been developed using stem-loop RT followed by TaqMan PCR analysis. Stem-loop RT primers are better than conventional ones in terms of RT efficiency and specificity. TaqMan miRNA assays are specific for mature miRNAs and discriminate among related miRNAs that differ by as little as one nucleotide. Furthermore, they are not affected by genomic DNA contamination. Precise quantification is achieved routinely with as little as 25 pg of total RNA for most miRNAs. In fact, the high sensitivity, specificity and precision of this method allows for direct analysis of a single cell without nucleic acid purification. Like standard TaqMan gene expression assays, TaqMan miRNA assays exhibit a dynamic range of seven orders of magnitude. Quantification of five miRNAs in seven mouse tissues showed variation from less than 10 to more than 30,000 copies per cell. This method enables fast, accurate and sensitive miRNA expression profiling and can identify and monitor potential biomarkers specific to tissues or diseases. Stem-loop RT-PCR can be used for the quantification of other small RNA molecules such as short interfering RNAs (siRNAs). Furthermore, the concept of stem-loop RT primer design could be applied in small RNA cloning and multiplex assays for better specificity and efficiency.
Figures



References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

